Instructions and Help
WebFR3D Help
This document contains information about search options implemented in WebFR3D. This separate tutorial describes how to set up WebFR3D searches. If you can’t find an answer to your question, please contact us.
Changes as of 2023-01-26
- Some superpositions were failing. One reason is because some nucleotides lack the P atom. Now we use C1' atoms for superposition when there are three or more nucleotides, and when there are two nucleotides, we use C1' and C3' atoms.
- See the article "Lone pair pi contacts and structure signatures of r(UNCG) tetraloops, Z-turns, and Z-steps: A WebFR3D Survey" by Zirbel and Auffinger (2022) for information about oxygen stacking interactions and for examples of WebFR3D searches and how to construct them.
Changes as of 2022-12-22
- On WebFR3D output pages, click on column headings to sort the table by that column. One can also sort by the checkboxes in the Show column. Clicking on the first column header, labeled S. for similarity order, restores the original order, which is the order in the heat map.
- Sorting does not change the order of the rows and columns of the heat map, but Next and Previous refer to the current sort order.
- Interactivity between the heat map, the table, and the coordinate window has been enhanced.
- Click on the diagonal of the heat map to select and view a single instance.
- Click below the diagonal to select and compare two instances.
- Click above the diagonal to select and view a range of instances.
- Right click or control click to add or remove from the current selection
- The selected instances are marked with a white square on the diagonal of the heat map
- Back end work was done to speed up the processing of geometric and mixed queries by processing the pairwise distances between units more quickly. Even so, geometric searches with a discrepancy cutoff above 0.5 and several positions may not return results; use symbolic constraints or a lower discrepancy cutoff.
Changes as of 2022-06-24
- The default specifier for unit type is "N" which expands to RNA nucleotides A, C, G, or U. To allow modified RNA nucleotides, type "RNA" in the corresponding white box. To require a modified RNA nucleotide, type "RNA modified" in the white box.
- Modified nucleotides can be queried using geometric searches and using chain continuity constraints, but no basepairs, base stacks, base-phosphate, or base-backbone interactions are annotated yet for modified nucleotides.
- We use PDB nomenclature for modified nucleotides, for example, 5MU instead of m5U.
- Not all modified nucleotides are available yet, just the most common modified RNA nucleotides. The available modified nucleotides can be seen on Github.
Changes as of 2022-05-21
- You can search representative NMR structures by choosing a representative set and then NMR in the resolution dropdown.
- Output includes a Sequence column which tells the sequence of the residues matching each position. Between the residues are the following symbols:
- - when the residues are successive in the chain in the usual order in the listing on that row
- → when the resides are in the same chain in the usual order but not successive
- ← when the residues are in the same chain but not in the usual order
- . when the residues are in different chains, or different symmetry operations on the same chain
- - when the residues are successive in the chain in the usual order in the listing on that row
- Chain direction constraints can be specified using "after" instead of ">" and "before instead of "<". "next" is an abbreviation for "=1 >" and "previous" is an abbreviation for "=1 <".
- Candidates will not include residues from different models
- Distance constraints may be separated by semicolons or spaces, for example, "=1;>" for sequentially adjacent nucleotides in increasing nucleotide order. Exact distance constraints are specified by separating possible distances by commas, for example, "=4,5" for distance of 4 or 5 in either direction, or "=4,5 >" for distance 4 or 5 and in increasing nucleotide order.
- Type "syn" or "anti" or "int_syn" in the white diagonal boxes for those glycosidic bond conformations. "is" can be used as an abbreviation for "int_syn". Multiple specifications are interpreted as "or". Negations work as expected; "~anti" is a synonym for "syn int_syn". These annotations are only shown in the results when you ask for them. You can force them to appear by typing "glyco" or "glycosidic" in one of the white diagonal boxes; this does not impose any constraint on the nucleotides.
- Glycosidic bond conformations map to chi angle this way:
- syn for chi from -45 degrees to 90 degrees
- int_syn for chi from -90 degrees to -45 degrees
- anti for chi from -180 degrees to -45 degrees or 90 degrees to 180 degrees
- The chi angle can be constrained to be between 100 and 120 degrees by typing chi(100:120) or chi_100_120 in the white diagonal box. The chi angle can be constrained to be above 170 degrees or below -170 degrees by typing chi(170:-170) or chi_170_-170.
- Note that constraints on basepairs (like tSH) and base stacking (like s35) need to go in the yellow boxes. They can be written in the opposite order (like tHS and s53) so that you can find an appropriate box to put them in. On the other hand, base-backbone interactions (like BPh and BR) cannot be written in opposite order, so you can specify those in either the yellow boxes or blue boxes.
- When giving multiple interaction constraints in the yellow boxes, they are interpreted as having logical "or" between them, unless the keyword "and" is used. Use that between groups of different types of mutually exclusive constraints.
- cWW cWH cWS ... will be interpreted as cWW or cWH or cWS, which makes sense because these are mutually exclusive
- tHH BPh ... will be interpreted as tHH or BPh
- tHH and BPh ... will be interpreted as tHH and BPh
- Nucleotides and amino acids are listed using unit ids, which are explained on the page about unit ids
Synopsis
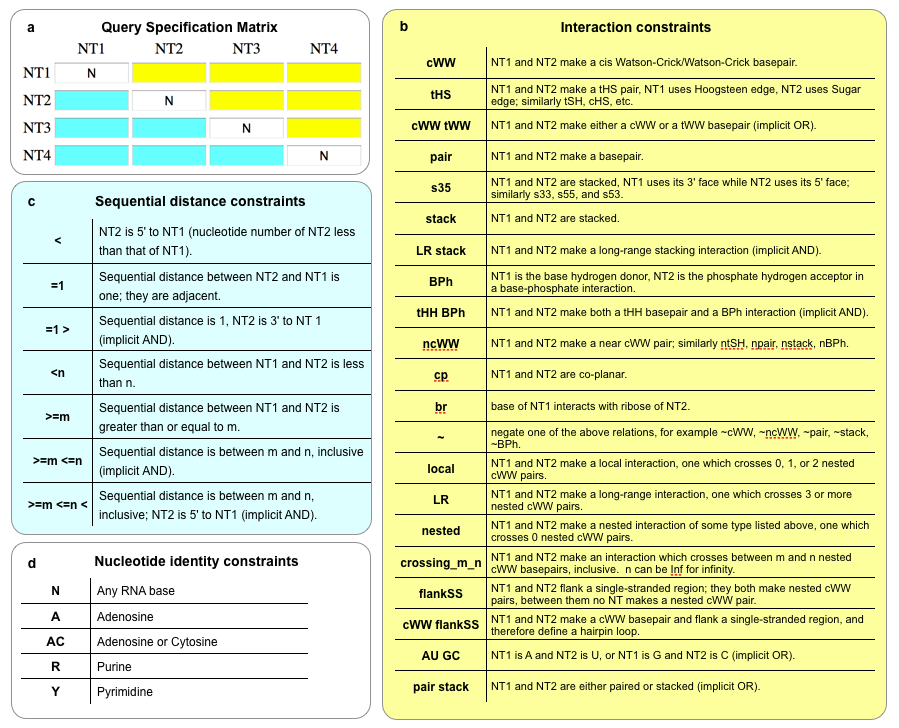
This diagram summarizes most of the available search options that can be entered in the Query Specification Matrix on the search webpages. Detailed descriptions can be found in the text below.
Sequential Distance Constraints
Set limits on the difference between nucleotide numbers using the boxes below the diagonal. (Actually, what is used is the difference between the index of nucleotides in the file, not NDB nucleotide number.)
To put an upper limit on the difference, type something like <5 or <=5.
To put a lower limit on the difference, type something like >5 or >=5.
To put both limits at once, type something like >5 <=12.
- To insist that the nucleotide in the given row have a lower nucleotide number than the nucleotide in the given column, type <, separated by a space from other specifications. For greater, type >.
Interaction constraints
Basepair, base stacking, or letter pair constraints are specified above the diagonal. To specify that all candidate motifs must have a tWH basepair between the nucleotides corresponding to the first and second nucleotides in the query motif, type tWH in the first row, second column. This means that the nucleotide in the first row must use its Watson-Crick edge, and the nucleotide in the second column must use its Hoogsteen edge. Base phosphate and base ribose constraints can be included either above or below the diagonal.
Valid basepair specifications are: cWW, tWW, cWH, cHW, tWH, tHW, cWS, cSW, tWS, tSW, cHH, tHH, cHS, cSH, tHS, tSH, cSS, tSS. Note, however, that the cSS and tSS interactions are not, in fact, symmetric, because each base can use the sugar edge differently. Following Leontis, Stombaugh, Westhof (NAR 2002), type cSs to specify that the first base has priority, csS for the second, or cSS for either. (Note: this feature is not currently enabled as of October 2021.)
Specifying multiple interactions allows more ways a candidate can satisfy the constraints; for example, typing cWH cHW requires a cis Watson-Crick/Hoogsteen basepair, but either base can use the Watson-Crick edge, and the other uses the Hoogsteen edge.
The abbreviation trans gives all trans categories, cis for cis.
Type bif for bifurcated basepairs (see NAR 2002).
Type ~cWW to exclude candidates having a cWW basepair.
Some pairs of bases are close to, say, cWW, but do not meet the strict criteria for membership in the cWW classification. Type ncWW (“near cWW”) to get basepairs that are not classified into any category, but for which the cWW category is the closest match, up to a certain fairly generous limit. Type cWW ncWW to get cWW and near cWW pairs, cWW. Type ntrans to get all pairs nearest to a trans pair.
Type s35 for base stacking in which the first base uses its 3 face, and the second base uses its 5 face. Similarly, type s53, s33, ors55. Type stack to allow all stacking interactions. The prefixes “n” and “~” work with stacking, as above.
To specify that the nucleotides must match a certain pattern, type, for example, cWW CG GC to get only CG or GC cWW pairs.
To require that two nucleotides make a base-phosphate interaction, enter BPh in the corresponding yellow box. This will select pairs of nucleotides in which the first nucleotide’s base is a hydrogen bond donor and the second nucleotide’s phosphate is an acceptor. To specify particular base-phosphate categories, type 0BPh, 1BPh, 2BPh, ..., 9BPh. For near base-phosphate interactions, type nBPh, n1BPh, etc. See the original paper about classification of base-phosphate interactions for more information.
Oxygen stacking is when a backbone oxygen of one nucleotide stacks on the face of the base of a different nucleotide. These can be specified with a constraint in the form s[oxygen][face] where [oxygen] can be O2', O3', O4', O5', OP1, OP2, and [face] can be 3 or 5. For example, sO4'3. Make sure to use a plain apostrophe character, not a "smart" one as word processing programs use. The constraint is directional and can be placed in either the yellow or blue boxes. s[face][oxygen] can also be used. Abbreviations can be used; sO3 is an abbreviation for sO2'3 sO3'3 sO4'3 sO5'3 sOP13 sOP23, and sO3' is an abbreviation for sO3'3 sO3'5, and sO is an abbreviation for sO3 sO5. Valid abbreviations are sO3, sO5, sO2', sO3', sO4', sO5', sOP1, sOP2, s3O, s5O, and sO.
One can restrict to pairs that play a certain role in the secondary and tertiary structure. For pairs that are nested, type “N” or “nested". For pairs that cross nested interactions but involve nucleotides in the same branch of the RNA, type local or “L”. For long-range or distant interactions, between different branches of the RNA, type long-range, distant, “D”, or “LR”. Note that “nested”, “local”, and “distant” are mutually exclusive. They can be negated with ~, but ~local only returns distant interactions, not nested ones.
Currently not enabled: To find bases which are in the same plane and are close enough that they may hydrogen bond in some way, type coplanar or cp. Near and not coplanar can be obtained with the "n" and "~" prefixes, respectively.
To specify bases that participate in cWW pairs and that delimit a single-stranded region such as a hairpin loop or one strand in an internal or junction loop, type "bSS" or "borderSS" or "flankss" or "flank". Note: for internal and junction loops, flanking nucleotides will be on the same strand, one on each side of the loop. Such flanking nucleotides usually do not interact with one another. In a hairpin, however, the nucleotides in the closing basepair simultaneously make a cWW pair and satisfy the borderSS relation.
Nucleotide identity constraints
The user can impose a nucleotide identify constraint (nucleotide mask) for their search by putting in nucleotide constraints in the text-boxes on the diagonal in the Interaction Matrix, which has a white background. Typing A, for instance, means that only candidate motifs with an A in the corresponding position will be kept. Typing AG allows either A or G, etc.
The program uses these standard abbreviations for other combinations:
- M for A or C
- R for A or G
- W for A or U
- S for C or G
- Y for C or U
- K for G or U
- V for A, C, or G
- H for A, C, or U
- D for A, G, or U
- B for C, G, or U
- N for A, C, G, or U
Note that N is the default. One may also exclude a given base using the syntax ~G
for instance, to exclude candidates with a G in the corresponding position.
Nucleotide numbers
Nucleotides must be separated by commas in the input page. Use unit ids as described on the unit id page.
RNA-containing PDB files
The list of PDB files is updated weekly (on Wednesdays) to include all available RNA-containing PDB files. WebFR3D also includes several representative sets of PDB files at various resolutions. More information about the representative sets can be found on the representative set website and in the NAR 2009 paper.
Discrepancy
Geometric discrepancy is a measure of how similar RNA structures are. Higher geometric discrepancy corresponds to more dissimilar structures. Identical structures have discrepancy zero. Searches with high geometric discrepancy cutoffs take significantly longer than those with lower cutoffs.
Geometric discrepancy is an entirely geometric measure that takes into account the general shape of the candidate motif and the orientations of its bases. First, we determine the shift vector and rotation matrix which map the geometric centers of the bases of each candidate motif onto the corresponding base centers in the query motif with the smallest error, called the fitting error. After the rigid body operations are performed, we compute the angles of rotation needed to align each base of the candidate with the corresponding base of the query motif. The square root of the sum of the squares (RMS sum) of these angles (in radians) is called the orientation error. The geometric discrepancy is defined to be the RMS sum of the fitting and orientation errors, divided by the number of bases in the query motif.
For more information about geometric discrepancy, please see the original FR3D paper.
Output columns
Here is a description of the output columns from left to right. Column order is subject to change as we refine the output. Columns with no data are suppressed.
- S is the candidate number. The candidates are ordered by similarity, S stands for Similarity.
- Show has a checkbox to display or not display the candidate in the coordinate window.
- Discrepancy appears in mixed and geometric searches. It shows the geometric discrepancy between the candidate and the query motif. As there is no query motif in a symbolic search, this column does not appear in symbolic searches.
- Resolution column tells the reported resolution of the structure in Angstroms. NMR structures will have 'NMR' listed instead.
- Position columns give the unit ids that match the positions in the query. There is a one to one correspondence between the query positions and the units in the candidates.
- Sequence gives the monomer type separated by symbols that tell the chain connectivity between the units. Units that are adjacent and in the order are separated by -. Units that are in the usual order are separated by a right arrow. Units that are in the same chain but not in the usual order are separated by a left arrow. Units that are in different chains or different symmetry operations are separated by a period.
- Orient tells the glycosidic bond orientation of nucleic acids. These will be printed when requested by typing "glyco" or "glycosidic" in a white search box or when using an anti, syn, or intermediate syn constraint.
- Chi tells the glycosidic bond angle of nucleic acids. It is shown when glycosidic bond orientations are shown.
- Columns with numerical headings like 1--2 show annotated pairwise interactions between units in the corresponding positions. Columns with no interactions are suppressed in the output to conserve space.
- Basepairs and base stacking come first
- Base-phosphate interactions come next
- Base-ribose interactions come next
- Crossing tells the number of nested Watson-Crick basepairs that the interaction crosses, and so measures how long-range the interaction is in a nucleic acid secondary structure. As of 5/6/2022, the crossing number includes all base combinations making a cis Watson-Crick / Watson-Crick basepair. In the future, it will only include base combinations AU, GC, or GU, including situations in which one or both bases are modified.
Ordering of instances
Instances matching the query are ordered by similarity, so that the more geometrically similar instances are placed near each other in the ordering. The methodology used is the "tree-penalized Path Length" or "tpPL" methodology of Aliyev and Zirbel (2022).
Heat map
The heat map displays the all-against-all geometric discrepancy between instances, with the instances listed in similarity order.
Clicking the diagonal of the heat map selects one instance to display in the coordinate window. Clicking a square of the heat map below the diagonal selects two instances (one according to the row, one according to the column) and displays their superposition in the coordinate window. Clicking a square of the heat map above the diagonal selects all instances from the row to the column, and displays these instances in the coordinate window.
You can optionally specify your email to receive a notification once your search has completed.
Updated: 01/26/2023 03:54PM
