Ribostral: An RNA 3D alignment
analyzer and viewer based on basepair isostericities
Table of Contents:
1. Introduction
Ribostral (Ribonucleic Structural Aligner) is a suite of programs designed
to integrate known structural data with homologous sequence alignments, with
the purpose of evaluating the quality of the alignments and guiding efforts
to improve them. The main GUI (Graphical User Interface) of this program provides
an expandable user-friendly platform through which other related programs can
be run. The related programs gather and analyze atomic resolution structure
data, parse and automatically align sequences, and perform other manipulations
on sequence alignments including extracting sub-alignments corresponding
to individual motifs or domains, removing
repeated sequences from an alignment to build a “unique” alignment
with higher phylogenetic diversity, and creating a .fasta alignment file from
.mat, which is another alignment format used by Ribostral. These tools are
covered in detail at the end of this manual. The main functions of the program,
namely analyzing, evaluating, and viewing RNA sequence alignments are discussed
first.
RNA sequence analysis programs are common, but none of them
provides the valuable structural information provided by ribostral. One of
the most widely used editors
for manual alignment of RNA sequences is the program BioEdit, which runs under
Windows platforms (1). BioEdit reads a sequence alignment file and
allows the user to choose pairs of nucleotides one pair at a time and display
substitution (covariation) patterns for them (in BioEdit, this is called Mutual
Information Examination). The resulting substitution table, however, provides
no description of what the observed substitution patterns mean in terms of
structure. BioEdit does provide one kind of link between sequence alignments
and structure: if the sequence alignment contains a “mask” describing
cis Watson-Crick nested basepair, BioEdit will color nucleotides in the alignment
in terms of how well they conform to the basepair family. A mask is a representation
of the locations of basepairs occurring between nucleotides represented in
the sequence alignment, with paired characters such as "(" and ")" indicating
the positions of basepairs in the alignment. BioEdit however does not provide
such information for any of the other eleven or so families of edge-to-edge
interactions that are possible between basepairs, comprising about one third
of all interactions (2). BioEdit is designed to be primarily
an alignment editor and viewer, and not a tool for structural alignment of
RNA the way Ribostral is. Coseq, a program that runs under UNIX platforms,
measures substitution patterns of basepairs without using any structural information
(Massire and Westhof, unpublished). The user then needs to analyze the
structure manually to see if sequence substitution patterns agree with it.
Finally, S2S is another more recent UNIX program that dynamically displays
parts of structure, creates full 2D annotations of them, and shows corresponding
positions in sequence alignments in an alignment editor (3).
However, it too does not evaluate alignments based on isostericities of basepairs
formed in structure the way Ribostral does.
Ribostral, which runs under multiple platforms, can either
be used like other sequence analysis programs, i.e., to simply provide substitution
patterns
of basepairs in a sequence alignment without any relation to structure,
or can
be used as the much more powerful tool that it is designed to be: to provide
substitution patterns, and at the same time superimpose structural information
on top of the substitution patterns to make sense of it. Ribostral does that
by coloring substitution patterns of each basepair in a way that reflects
the edge-to-edge family and the isosteric subfamily it belongs to (4).
Even in its more simple usage as a program that provides sequence substitution
analysis without the use of 3D structural information, Ribostral
is more convenient
than other programs, because it allows for simultaneous analysis of lists
of basepairs and produces a single and easily portable HTML output, without
having
to input nucleotide numbers of interest one position at a time. Interactive
position-by-position analysis is also possible, where in addition to what
has been described above, an integrated structure viewer is also available.
In
addition to providing substitution information for basepairs, Ribostral is
also capable of analyzing substitution patterns for more than two nucleotides
at once, such as base triples, quadruples, and so on. The sequences of whole
motifs can be analyzed this way, as will be shown below.
1.1. Supported platforms and deployment process
Ribostral is designed and fully tested under Windows XP. It is free and can
be obtained from http://rna.bgsu.edu/Ribostral. The program is distributed
in two forms: MATLAB source files capable of running on any PC or MAC platform
with MATLAB version 7 SP3 or higher (with loss of some non-essential options
on
MAC), or a stand-alone program capable of running under the PC
platform, after installation of a free compiler provided by Mathworks
(details can be found here).
Figures used in this text are based on the Windows XP version with system
appearance set to Windows Classic style. Upon downloading the MATLAB source
files or the stand-alone version, the user will end up with the specific
hierarchy of folders shown in Figure 1.
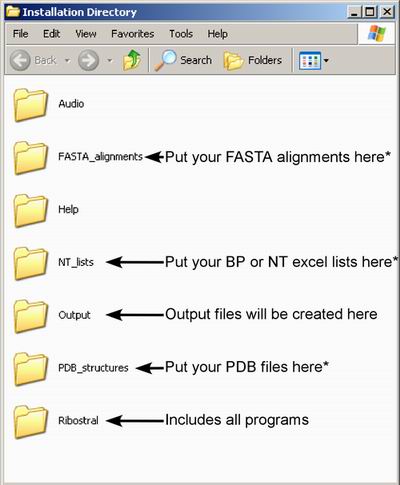
Figure 1. Ribostral default installation subdirectories. Using
folders marked with asterisk (*) is optional; on Windows platforms, these are
the locations where Ribostral starts browsing for the corresponding input files.
1.2. Disclaimer
No guarantee, expressed or implied, is made as
to the suitability of this software for any purpose, computer, or person. The
author shall not be held responsible, nor be liable for any damage occurring
in any way to equipment or health while using this software.
Author contact information: Ali Mokdad, M.D., Ph.D.,
Department of Biological Sciences, Bowling Green State University, Bowling
Green, Ohio 43403. Email:
mali@bgsu.edu.
2. Executing
Ribostral
Ribostral can run as a GUI or as a script under MATLAB (by
running the program ribostralNoGUI; follow the header information to point
to your files of interest). The following discussion concerns mainly the
default GUI version.
The startup window of Ribostral is a blank GUI with
three menu options: File, Tools,
and Help
(Figure
2).
The
other
GUIs
of the
program
are activated
from
this window.
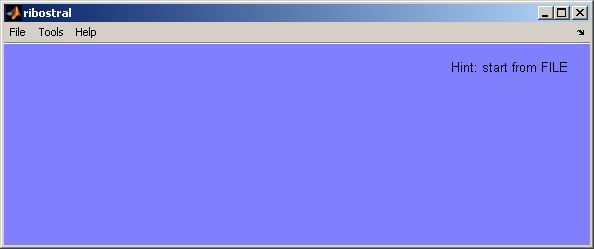
Figure 2. Ribostral’s main GUI.
To avoid ambiguity, the GUI only displays options that are
allowed at the specific stage of the analysis. For example, before doing any
analysis the user needs to load a sequence alignment file on which the analysis
will be performed. So initially, the GUI does not display the buttons that
start the analysis. These buttons will appear only after the sequence alignment
file is loaded into the program. To provide additional help for the user, the
upper right-hand corner of the main GUI is reserved for messages describing
the status of the program, any errors in execution, or hints on how to proceed
further.
2.1. Loading an alignment file
To start the analysis, the user needs to follow the hint that appears in the
upper right-hand corner of the main GUI: “Hint: start from FILE”.
By clicking on “File” from the menu bar, the user sees the options:
Open FASTA alignment, Open NT list, and Preferences (Figure 3).
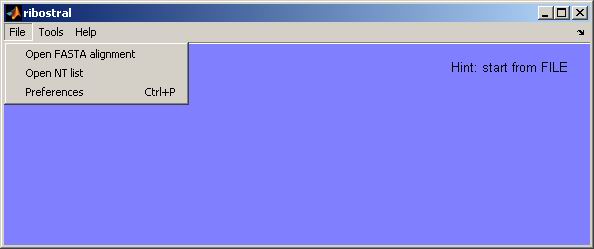
Figure 3. File menu options.
The first thing to do is to open an alignment file. In its
current version, Ribostral reads only alignments in FASTA format, the simplest
and most common sequence alignment format available. In this format, each sequence
is represented by a header line preceded by the symbol “>” and
one or more sequence lines. Upon choosing “Open FASTA alignment” from
the File menu, the user can browse the local drive for alignment files (Figure
4).
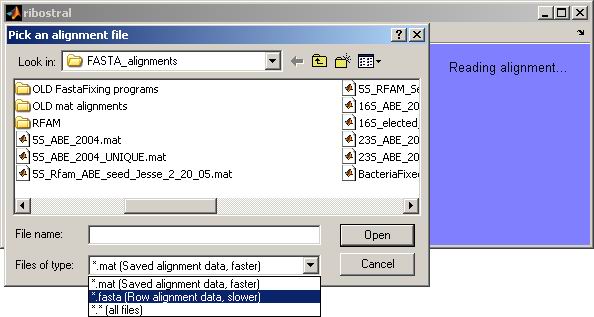
Figure 4. Browsing for an alignment file.
On PC machines, the browser window initially starts looking
for alignment files in the folder:
“<
installation directory>\FASTA_alignments” (refer to Figure 1). Here,
there are two main options: reading a raw alignment file in its text form (with
extensions such as .fasta, or .txt), or reading a MATLAB data file (.mat) which
is derived from the raw alignment file. This last option is processed faster
by Ribostral, especially if the alignment of interest contains thousands of
sequences, like the 16S rRNA alignments. A .mat file is created after a FASTA
alignment is read for the first time. It is saved in the same directory and
under the same name except for the extension. Time can then be saved by reading
the data file instead of the text file the next time the same alignment is
used. Note that a new data file is only created if no data file with the same
name is already present in the directory. Therefore, if the raw FASTA alignment
is deliberately modified in any way, the old .mat file referring to it needs
to be deleted to allow for its re-creation. Notice that when a FASTA format
file is read, all characters indicating unknown nucleotides (“N”, “n”, “O”,
and “o”) are transformed into “o”, and all other characters
are capitalized. When analyzing basepairs, Ribostral only recognizes dashes
(“–”) which represent gaps or deletions, o’s, and the
four RNA nucleotide letters A, C, G, and U as valid characters in sequences.
When analyzing longer motifs, the program recognizes all characters.
2.2.
Dividing Sequences into Subgroups
After choosing the alignment file of interest, Ribostral looks in the same
directory for an Excel file called “KnownFASTAFilenames.xls”. This
is an optional user-created Excel sheet that gives additional details about
the FASTA file being read, such as the names of different groups (or domains)
of organisms it represents, and the boundaries of these groups. If this Excel
file is not present in the same folder as the FASTA file being read, or if
the exact name of the FASTA file being read cannot be found in the Excel file,
this information will be ignored and all FASTA sequences in the alignment will
be considered as one group. The analysis of specific subgroups (such as phylogenetic
domains) will not be possible later. Figure 5 shows the format of the “KnownFASTAFilenames.xls” file
and the information it contains.
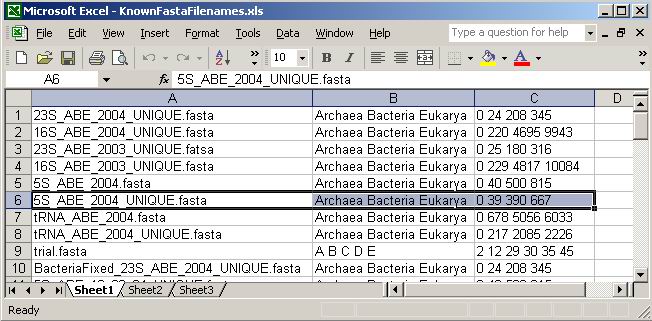
Figure 5. A snapshot of the “KnownFASTAFilenames.xls” file.
The highlighted entry (number 6) is the FASTA file analyzed in this manual.
The Excel file is organized in the following way: The first
column contains the exact names of “known” FASTA alignments; the
second column contains the names of the subgroups the FASTA alignment contains
separated by empty spaces; and the third column defines the limits of these
subgroups in the order of their names, separated by empty spaces. The highlighted
file (entry 6, 5S_ABE_2004_UNIQUE.fasta) is the one used for the sample study
in this manual. It contains sequences from the phylogenetic subgroups: archaea
(sequences 0+1 to 39), bacteria (sequences 39+1 to 390), and eukarya (sequences
390+1 to 667). To ignore the first sequence in the alignment (if it is a structure
mask for example, or if it is a reference sequence that does not belong to
the subgroup), “1” can be placed instead of “0” in
the third column. In that case, the program still reads the first sequence
but does not include it in the analysis.
The main GUI then displays any known details associated with the FASTA file
and also unlocks some buttons or check boxes that permit the user to analyze
the alignment (Figure 6). Note that if this step (loading a new FASTA file)
is repeated at any stage during the analysis, all previous data that corresponds
to previously read alignments will be erased from memory and the GUI will be
reinitialized.

Figure 6. GUI changes when an alignment file
is successfully loaded. The text box in the middle displays the name of the
sequence file in memory.
At this stage, two options are possible for carrying out sequence
(and optionally structure) analysis: analyzing a list of nucleotides (with
or without structure data) for the complete sequence analysis of all positions
listed in it, or interactively and directly analyzing individual positions
on screen. The first option is covered first.
2.3.
Preparations for the analysis of a list of nucleotides
After loading a FASTA file, the user can load an Excel file containing a list
of the nucleotides of interest. This is done by activating the option “Open
NT list” from the File menu. A browser opens and on PC platforms search
for Excel files starts in the local directory “<installation directory>\NT_lists” (Figure
7).
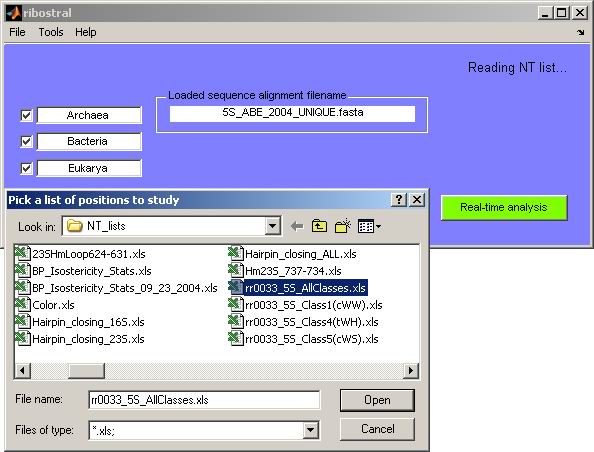
Figure 7. Browsing for an Excel nucleotide list.
The NT list Excel file must contain names of the source
organisms and nucleotide numbers of interest. The first row is a header row
and anything in it will not be read. Figures 13 and 14 show what such a file
should look like.
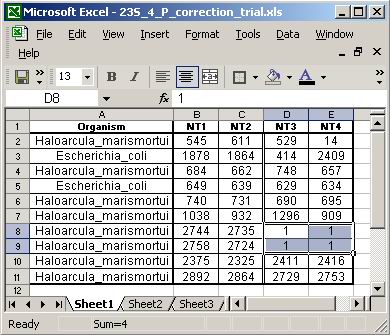
Figure 8. Nucleotide list for analysis. This
Excel list from 23S rRNA allows the sequence analysis of four nucleotide positions
simultaneously.

Figure 9. Basepair list for analysis. This
Excel list from 5S rRNA includes structural data about some of the basepairs
(to show that structural data is optional).
As seen in Figures 8 and 9, the first column of the Excel
sheet contains the name of the reference organism for that row, and subsequent
columns contain the nucleotide numbers to be analyzed. The name must be the
full name or part of a name (case insensitive) present in the loaded FASTA
alignment file. It does not necessarily need to be the beginning of the name
(e.g. “Halomari” can be used if the actual name in the FASTA alignment
is “Eu_halomari”). If several FASTA comment lines share this name,
the first of the occurrences will be considered as the reference organism (e.g.
if “halo” is used, and it is present three times in the sequence
alignment, first as “EU_HALOJAPO”, then as “Eu_halomari” and
then as “Eu_halomedi”, the first will be used). It is advisable
for the user to manually check sequence names in the FASTA file to prevent
inadvertent reference to unintended organisms. The most common error is caused
by incorrect spelling of the source organism in the NT list Excel file; if
such a spelling cannot be found in the FASTA file an error message will appear
and the program will stop. If universal numbers for the alignment are desired
the word “universal” can be used instead of an organism name (similar
to entry 8 in Figure 9).
Structural information (i.e. basepair type) is allowed only
for basepair lists. These are lists comprising two columns of nucleotide numbers,
like
the one
shown in Figure 9. Basepair types are named by reference to the interacting
edges of the nucleotides. These are the Watson-Crick edge (W), Hoogsteen
edge (H), and sugar edge (S). Edge-to-edge interactions in addition may
be cis (c)
or trans (t) with respect to the glycosidic bond. This gives rise to twelve
main families of basepairs (2,4), and
some intermediate families (5), only one of which with
currently characterized isostericity matrix (the bifurcated cWW/tWH family).
These thirteen
basepair types are coded as follows in order for Ribostral to understand
them (case insensitive for all except tSs, because it is a directional
interaction
with asymmetric isostericity matrix): cWW, tWW, cWH, tWH, cWS, tWS, cHH,
tHH, cHS, tHS, cSS, tSs, and bif. Asymmetric codes (such as cWS) can be
reversed,
so in the table if nucleotides 22 and 26 in this order are coded as tSW,
this is the same as 26 and 22 being tWS (see Table 1 for a definition of
all the
basepair codes used by Ribostral). Ribostral always presents data about basepairs
as one of the thirteen codes listed above (and not the reverse form).
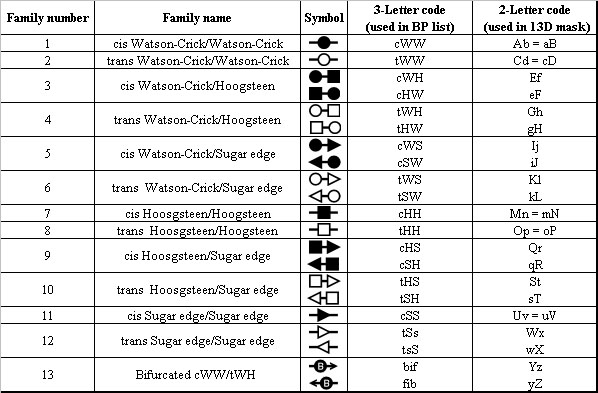
Table 1. Basepair codes used in Ribostral.
The last column shows the codes used for constructing the structural mask in
Alignment Viewer, discussed later in text.
If the type of basepairing is known for a pair of bases, the
output of the sequence analysis will be colored to indicate its isosteric subfamilies,
so the investigator can determine whether the aligned nucleotides for each
sequence represent isosteric or near-isosteric substitutions. If no structural
information is available, the table output will only display sequence substitution
data observed in the corresponding columns of the alignment.
After successfully reading a nucleotide list file, Ribostral displays a new
button allowing for the analysis of the whole list at once (Figure 10).
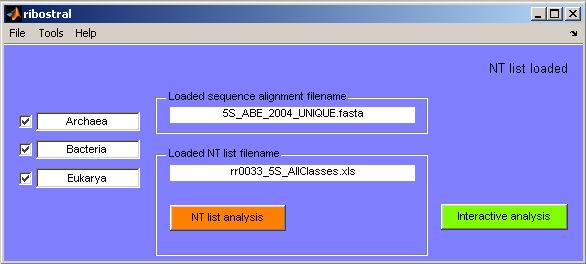
Figure 10. The main GUI after successful loading
of a nucleotide list file. The status bar (upper right hand corner) displays “NT
list loaded” and the new button “NT list analysis” becomes
available.
2.4.
Analyzing a list of nucleotides
Upon activation of the button “NT list analysis” the program takes
each row of the Excel NT list file and counts the number of each substitution
of its nucleotides in corresponding columns of the alignment. This process
may take a few minutes for long lists of nucleotides or for large alignments.
The MATLAB command prompt (or the DOS prompt if the compiled stand-alone version
of Ribostral is executed) displays different messages and counters indicating
the status and progress of the analysis. Upon successful execution, output
files are created in the folder “<installation directory>\Output” (refer
to Figure 1), with descriptive names indicating the NT list file and the alignment
file they represent. At the same time a new button providing a quick link to
the main output file will also appear on the main GUI.
3.
Types of output files and their interpretations
Depending on the type of input NT list, several types of output files are
possible (Figure 11). The following sections discuss each type of these output
files
in detail.
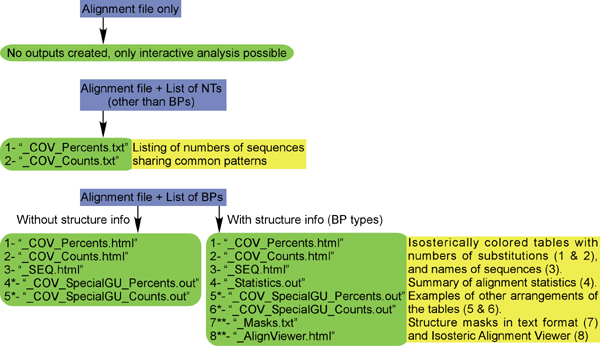
Figure 11. Output files produced depending
on input files analyzed. The first output file in each case is the “main” file
accessed directly by clicking the button “Display list results” on
the main GUI. Outputs indicated by asterisks (*) are produced optionally by
changing the program preferences.
3.1. Output
files for a non-basepair list
If the input NT list is not a list of basepairs but a list of nucleotides forming
distinct motids (such as the one shown in Figure 8), two text output files
will be created, one giving counts and the other percentages of sequences that
share a common pattern (Figure 12). The patterns are listed in alphabetical
order. To clarify this, the following example from Figure 12 is considered.
The first entry in the Figure (highlighted) shows that in the archaeal part
of the alignment, nucleotides corresponding to Haloarcula_marismortui local
numbers 545, 611, 529, and 14 (in this order) are 88% GUGC and 12% GUGo. From
this one can deduce that Nucleotides 545, 611, and 529 are GUG in 100% of the
cases, and so on. The values in the percent output file are rounded according
to the preferences of the user (Ribostral preferences will be discussed in
detail later).
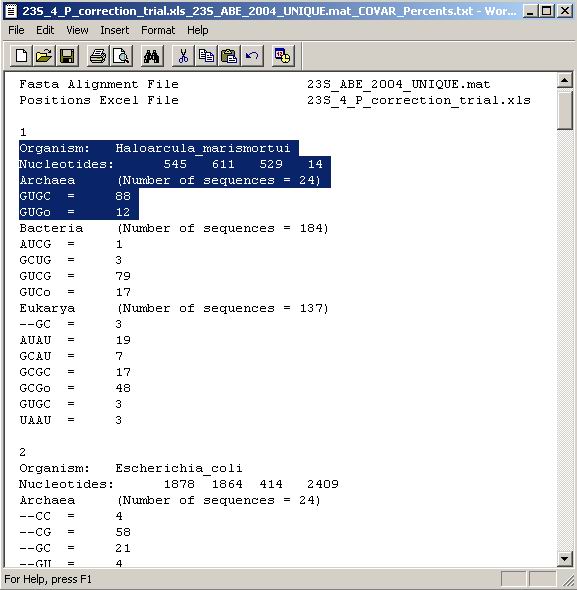
Figure 12. Percent output text file created
upon sequence analysis of an NT list. Sequence patterns are listed in alphabetical
order.
A similar output file with counts instead of percents is also created. This
analysis is for the NT list shown in Figure 8.
3.2.
Output files for a basepair list
In case the Excel input NT file gives a list of basepairs from an atomic resolution
structure, Ribostral generates output files that are better tuned and more
informative for this kind of input. These include colored HTML outputs in the
form of tables with sequence substitution values. The tables use different
background colors to indicate for each basepair whether observed substitutions
are isosteric, near-isosteric, heterosteric, or forbidden as compared to what
is observed in the structure. By default, three HTML output files (and a fourth
text output in case the input Excel list is a basepair list with structure
information) are produced upon the analysis of a basepair list (see Figure
11). The main output file accessed directly by the “Display list results” button
on Ribostral is the percent covariation file whose name ends with “_COV_Percents.html”.
A similar “_COV_Counts.html” file shows the counts instead of percents.
Figure 13 displays a snapshot of such an output. A third file (ending with “_SEQ.html”)
displays the names of organisms giving rise to the values in the first two
output files. The positions of the names correspond to the values observed,
as clarified in Figure 14. Note that the basepair identity observed in structure
is indicated by bold font in the corresponding cell.
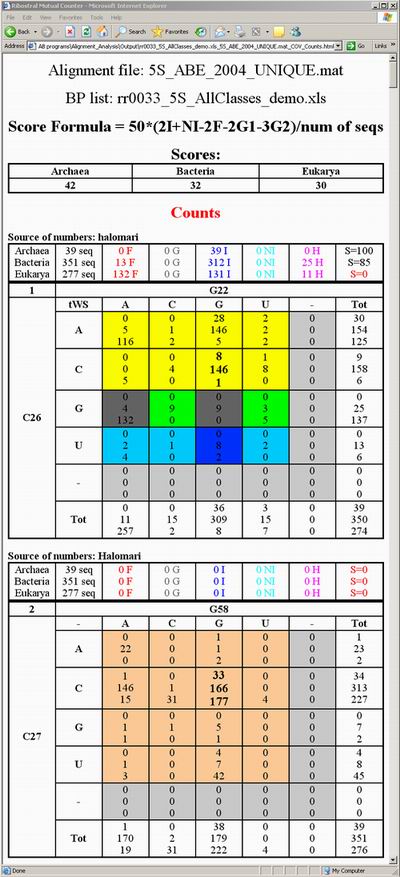
Figure 13. Percent output HTML file created
upon phylogenetic analysis of an basepair list. A similar output file with
counts instead of
percents is also
created. This analysis is for the NT list presented in Figure 8 above.
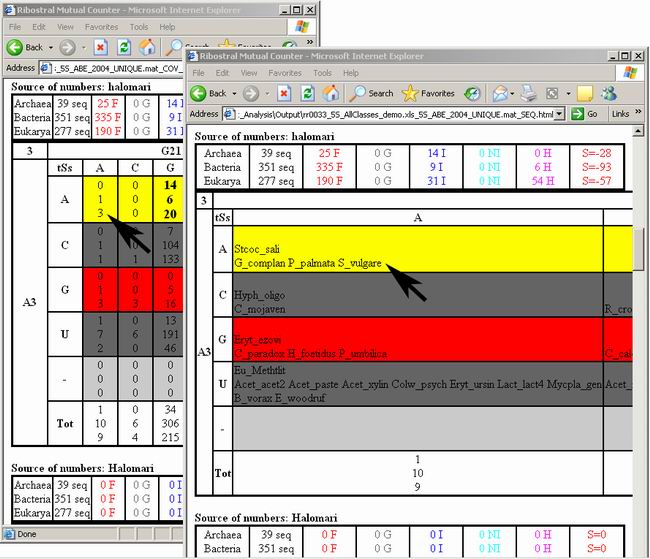
Figure 14. Matching sequence values and names.
The “_COV_Counts.html” output file (left panel) displays the sequence
counts, and the corresponding “_SEQ.html” (right panel) displays
the names of sequences. Note the value indicated by the black arrow in both
outputs: The left panel shows that there are three bacterial counts for AA
occupying the position A3/G21 (halomari local numbers). The right panel shows
that these three counts come from the sequences of G_complan, P_palmata, and
S_vulgare.
The output files have two purposes: First, to display substitution
patterns corresponding to each BP position analyzed, and second, to provide
information about how well each of these positions in alignments agrees with
3D structure. Based on this, if there are any potential mistakes in the alignment
they are easily pinpointed for their manual correction with available sequence
editors.
All three HTML output files mentioned start with a total score assigned for
each phylogenetic subgroup in the alignment (in this case the subgroups are
the three domains: archaea, bacteria, and eukarya). Each subgroup will have
its own results printed on a separate line in the substitution tables. The
areas in Figure 14 indicated by the black arrows for example represent the
substitutions in eukaryal sequences of the basepair A3/G21 (using halomari
local numbers). On top of each table that represents a basepair position the
following information is provided: The names of the subgroups in the order
they are analyzed, the numbers of sequences in each subgroup, the count of
forbidden substitutions present in the table (in red color, with the letter
F following the value), the count of gaps present including gaps on both sides
or just on one side (in gray, with the letter G following the value), the count
of isosteric substitutions (in blue, with the letter I following the value),
nearly-isosteric substitutions (in cyan, with the letters NI following the
value), non-isosteric but not forbidden substitutions that we refer to as heterosteric
substitutions (in pink, with the letter H following the value), and finally
individual scores of each subgroup at that basepair position.
The score is a value describing how structurally “acceptable” the
sequence substitutions at analyzed basepair positions are. This is based on
the measure of how structurally compatible these substitutions are with the
basepair in the reference organism, which is typically the organism with known
structure. The individual basepair scores are currently calculated based on
an ad hoc formula derived from experience with structures and knowledge of
the patterns of allowed substitutions for each type of basepair. This formula
can be easily modified by the user, by manually entering desired weight parameters
in the file “<installation directory>\Ribostral\SCORES.txt”.
The formula used throughout this text is:
Individual BP score = c * SUM(3I + 2NI – H – 2F – 2G1 – 3G2)
/ number of sequences
Where c is the correction coefficient: c = 100 / (Highest positive weight),
in this case c =100/3.
I, NI, H, F, G1 and G2 are the counts of sequences having substitutions that
are isosteric, nearly-isosteric, heterosteric, forbidden, gap on one side,
or gaps on both sides respectively. The Highest positive weight is 3 in this
case. The correction coefficient c insures that the maximum score is +100 (in
this case, the minimum score when all substitutions are gaps on both sides
is –
100, but that is not always the case depending on the formula used). The formula
used here is asymmetric: unfavorable terms that contribute to its reduction
are more numerous and weigh slightly more than favorable terms that contribute
to its increase. But for our purposes, this is not a critical point. What is
important is to easily identify low-scoring spots in the alignment to guide
manual realignment efforts. To better locate these trouble spots in the alignment,
the score is printed in red if it is worse than a certain threshold (i.e. if
it is below zero, which is the score in case no structural data is available)
and in black otherwise. The total score printed at the top of each HTML output
file is an adjusted sum (sum divided by number of basepairs studied) of the
individual BP scores in the study (so it is actually an average of the individual
scores). This means that its maximum is also +100 and its minimum, in case
the formula presented here is used, is –100.
Note that another valid formula used can be something like:
Individual BP score = c * SUM(2I + NI – 3F) / number of sequences
Both formulas are ad hoc and the difference between their weights is not essential.
The ease of reading and understanding normalized values between +100 and –100
or –150 is the reason for using a correction coefficient.
The most obvious and important element of the HTML output files discussed
thus far is the background color patterns that appear in their tables. These
colors reflect the isostericity matrices of the thirteen characterized families
of basepairs. Each family has its own pattern of isosteric, nearly-isosteric,
heterosteric, and forbidden substitutions. Boxes with the same colors are isosteric.
Boxes that are nearly-isosteric with each other have “similar” colors.
There are five groups of these “similar” colors: pink/red/orange,
red/orange/yellow (so substitutions with pink and yellow backgrounds are not
nearly isosteric to each other), blue/cyan, dark green/light green, and brown
(Figure 15). Forbidden boxes are colored in dark gray, and gaps in light gray.
Boxes with non-similar colors indicate that they are heterosteric to each other.
If the basepair family for a certain basepair is not specified, all nucleotide
boxes in its table will be colored in neutral beige (like the second BP listed
in Figure 13).
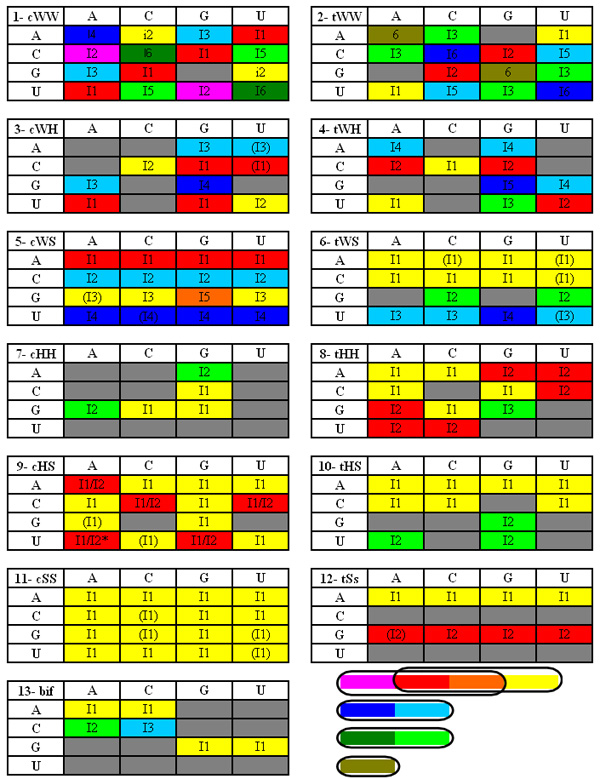
Figure 15. Basepair
families and their isosteric subfamilies. Gray colors indicate forbidden combinations
of nucleotides,
i.e., combinations
that cannot form basepairs due to steric clashes or incompatible distribution
of H-bond donor or acceptor atoms. In each family, isosteric subfamilies have
the same color, and nearly isosteric-families have similar colors (the five “similar” color
groups are shown on the bottom right corner). Letters correspond to the original
reference (4). Asterisk (*), corrected from the original
reference.
One more output file is produced by default in case a basepair
input list with structure information is analyzed. This is the “_Statistics.out” file,
which is best viewed with Microsoft Excel or WordPad. This output summarizes
some of the results by stating the percentage of basepairs analyzed that have
mostly allowed substitutions (containing <10% forbidden or gaps), the percent
of basepairs that have >10% forbidden substitutions, and the percent of
basepairs having >10% of sequences with gaps at their positions. It also
shows the percent of basepairs among the ones mostly allowed that have only
isosteric substitutions. Instead of the total score which is just one value
that describes the quality of the alignment studied, this output file gives
a more quantitative measure of the alignment quality.
When a Basepair list is analyzed, a button labeled “Plot scores” also
appears on the main GUI. When activated, this allows the user to quickly analyze
the scores of each basepair and define places of misalignment or motif swaps
(Figure 16).

Figure 16. Score plots for the archaeal
5S rRNA alignment. The red curve describes the scores of corresponding basepairs,
and the blue
line represents the average score (or total score as defined in this text)
for the whole alignment based on the basepair list provided. By modifying the
ad hoc formula according to which scores are calculated (this can be done by
changing parameters in the “SCORES.txt” file), the user can plot
any combination of one or more aspects of the alignment, such as percent isosteric
substitutions, percent isosteric and near-isosteric substitutions, and so on.
3.3. Ribostral Alignment
Viewer
There are additional output files not produced by default that can be obtained
by changing the Preferences under the File menu options. The reason for not
producing those output files by default is not to overwhelm the casual user
with too many output files, and not to unnecessarily extend the execution time.
If the “AlignViewer” option is selected in Preferences, an HTML-format
alignment viewer is created where the sequence alignment is shown in colors
indicating substitutions that are compatible with the 3D structure and those
that are not. This tool is the first of its kind taking into consideration
all thirteen families of basepairs instead of just the classical cis Watson-Crick
family. The sequences in the Alignment Viewer are colored in a way to describe
how well each column (more precisely, each pair of columns) in the alignment
agrees with structure (Figure 17). Basepairs from each sequence are colored
individually based on their isosteric agreement with the homologous basepair
from the reference sequence, which is usually the sequence with known 3D structure.
If the substitution pattern is isosteric to the one in the reference sequence
it is colored in green, if it is nearly-isosteric to it, it is colored blue,
heterosteric is pink, forbidden is red, gap on one side is dark gray, and gap
on both sides is light gray. If no basepair information is available the nucleotides
are printed in black (these color assignments can be modified by changing the
MATLAB script file “mColorCode.m”). For easy interpretation, the
color scale is printed at the top of the alignment viewer HTML file. Thanks
to this tool, it is now possible to look at the sequence alignment and directly
get a good idea about whether the alignment is structurally valid or not, and
where the major areas for local alignment mistakes (or motif swaps) are located.
Note that if base triplets are present (i.e., two basepairs with one nucleotide
in common), the basepair listed earlier in the Excel BP list file takes priority
in coloring. If the user prefers to give priority to cWW interactions for example,
then these should be listed first in the BP list.
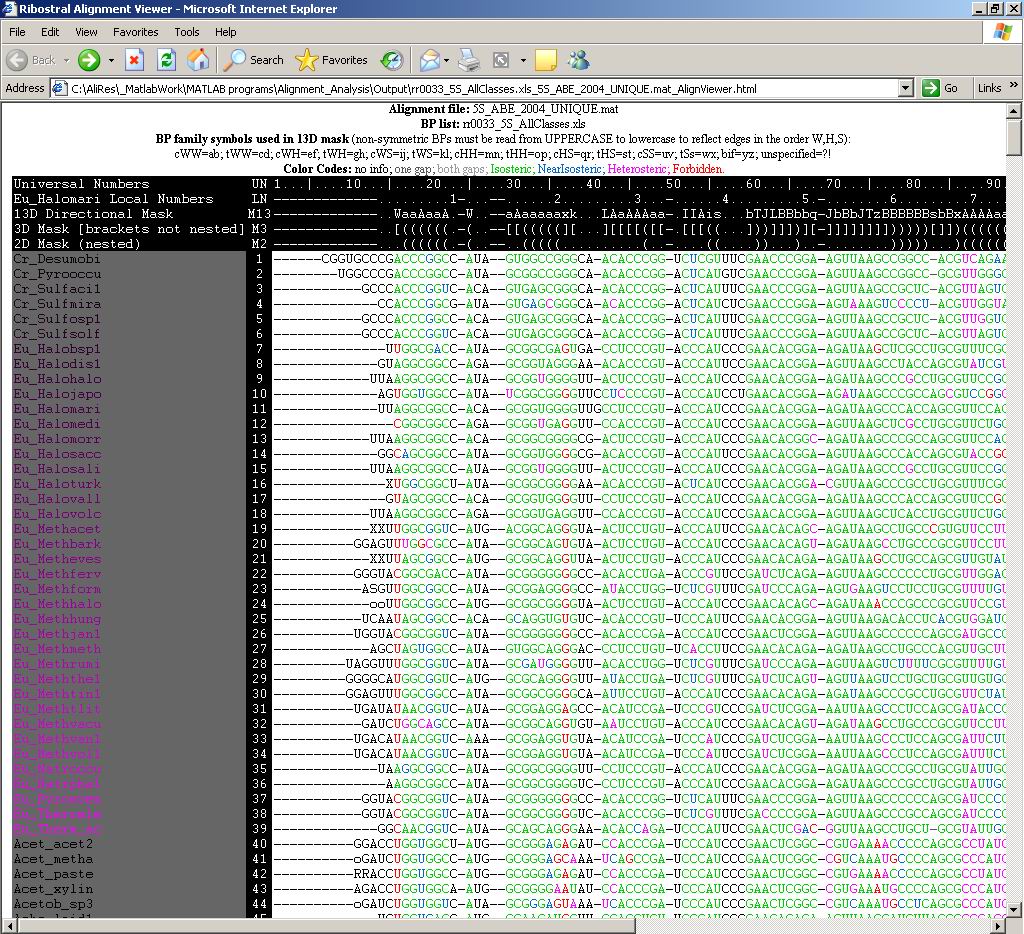 Figure
17. Ribostral HTML alignment viewer. The visible part of the 5S alignment
describes how well sequences agree with structure, represented here by Eu_Halomari,
or sequence number 11. The color code is printed at the top. Note that the
whole content including colors can be copy/pasted into Excel or other editors
for manipulation.
Figure
17. Ribostral HTML alignment viewer. The visible part of the 5S alignment
describes how well sequences agree with structure, represented here by Eu_Halomari,
or sequence number 11. The color code is printed at the top. Note that the
whole content including colors can be copy/pasted into Excel or other editors
for manipulation.
Ribostral Alignment Viewer starts with several title lines
containing the names of the analyzed files as well as the legend of all codes
and colors used. Following this is the listing of all organism names and their
sequences, exactly as they appear in the original FASTA alignment. Organism
names change their color gradually as they approach the limit of the subgroup
or domain they belong to. This is how it is made clear that sequence 39 for
example is the end of the archaea domain. The first five sequences (those on
black background) represent the universal numbers, local numbers, and three
types of structural masks that describe basepairing patterns. Universal numbers
are assigned to each character seen in the sequence, including all indels (“–”).
Local numbers are the numbers that correspond to the reference sequence; unlike
universal numbers, local numbers are not assigned to indels in this sequence.
In the Alignment Viewer only decimal representatives of local numbers are listed,
i.e. the first “1” stands for 10, the first “2” stands
for 20, and so on until the first “0” is seen, which stands for
100. After that the numbering cycle is repeated, so the second “1” stands
for 110, the second “2” stands for 120, and so on.
The structural masks describe basepairing patterns reported in the Excel BP
list directly on top of the sequences. Figure 18 is a schematic description
of the information represented by structural masks.

Figure 18. Schematic explanation
of structural masks. These are one dimensional representations of structure.
The example shown here is from Helix 95 containing the sarcin/ricin motif
in the large ribosomal subunit of H. marismortui (pdb file 1S72). Color codes
correspond to basepairs. Note that the 13D and 3D masks may have several
symbols overwriting each other; only one of the cyan RG and ][ symbols displayed
in this Figure on top of each other is displayed in the 13D and 3D masks
produced by Ribostral.
The 2D mask describes only nested basepairings, irrespective
of their geometric families (most of these would be helical cWW basepairs).
Each nested basepair (one which does not cross in 2D with other basepairs)
is represented by the two parentheses symbols “(” for its opening
or 5’ nucleotide and “)” for its closing or 3’ nucleotide.
The 3D mask is the same as the 2D mask, but in addition to representing nested
basepairs it also represents other non-nested basepairs using the bracket symbols “[” and “]”.
When one nucleotide forms more than one basepair by using more than one of
its edges at once (e.g. in base triplets), the number of bracket symbols of
the opening and closing type may not be equal. Therefore, it is not possible
from this mask alone to know which opening brackets correspond to which closing
brackets. Another limitation of both the 2D and 3D masks is that they do not
tell anything about the type of basepairs formed. The 13D mask is designed
to solve this problem. Here, instead of just one or two pairs of symbols representing
opening and closing of basepairs, thirteen different pairs of symbols are used
to represent all known families. These codes are AB for the cWW family, CD
for tWW family, and so on (see Table 1). Since some families also are not symmetric
(C/G cWS has a different meaning than C/G cSW), uppercase symbols and lowercase
symbols are used to indicate the direction of the interaction. The symbols
must be read from uppercase to lowercase direction whenever the interaction
is asymmetric, with the uppercase symbol referring to the base edge of higher
priority (edge priority is assigned in the order W, H, S). Thus, if an interaction
exists between columns (universal numbers) 38/42 and is represented by kL,
this means that nucleotides occupying these positions form a tSW interaction
in the source organism (or structure). This is the same as saying that 42/38
is a tWS (or Lk) interaction. The 13D mask has the same limitation as the 3D
mask, in that finding matching opening and closing symbols is not always straight
forward. Using the 2D mask together with the other two masks makes it easier
to at least find nested pairs.
3.4. Other output formats
Ribostral can also create other output files upon request (by checking the
option “GU special” in Preferences). These files represent another
way of classifying some similar isosteric groups together, the way the G/U
wobble basepairs are analyzed in our previous work (6).
Ribostral scripts can easily be modified to produce other similar outputs.
4.
Interactive analysis of nucleotides
Besides analyzing lists of nucleotides or basepairs at once and producing dedicated
output files that describe their results, Ribostral is also capable of analyzing
nucleotides of interest one by one and directly displaying their results on
screen. This can be done by activating the button “Interactive analysis” from
the main GUI, which starts a new GUI window (Figure 19). This option becomes
possible only after successful loading of a FASTA alignment file. If a BP list
file is also loaded before the interactive analysis is activated, the new GUI
will have more options and information extracted from that file. In the following
text we will discuss the options available in case a BP list is loaded before
activating the interactive analysis GUI.
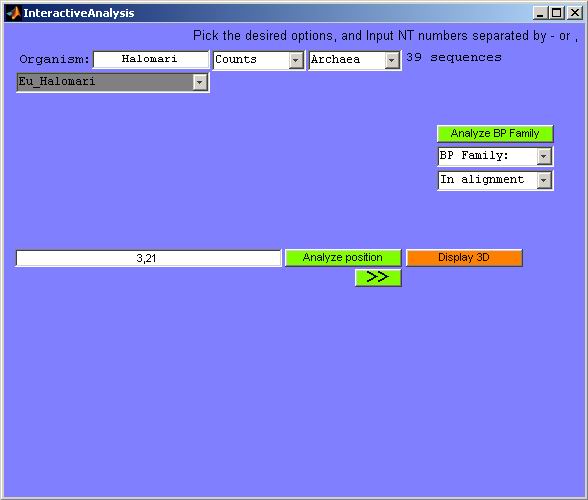
Figure 19. Initial screenshot of the interactive analysis GUI. Here, both
a FASTA file and an NT list file were loaded from Ribostral main GUI before
starting this GUI. If only a FASTA file was loaded, some of the options or
buttons seen here would not have been made visible.
The interactive analysis tool is a powerful tool providing
yet a new array of functions not provided by the list analysis tools discussed
in previous sections. After specifying the right choices (such as source organism
name, domain of interest, etc…) the user can either analyze a specific
family of basepairs, or analyze a particular position.
4.1.
Interactive analysis of a family of basepairs
The right-hand side of the interactive analysis GUI contains the options and
buttons capable of gathering statistics about a whole family of basepairs at
once. This option is only possible if the user has loaded a basepair list before
opening this GUI. An example of what can be done here is the analysis of all
the occurrences of the tHS family in the archaeal part of the 5S rRNA alignment.
The result of such an analysis is shown in Figure 20.
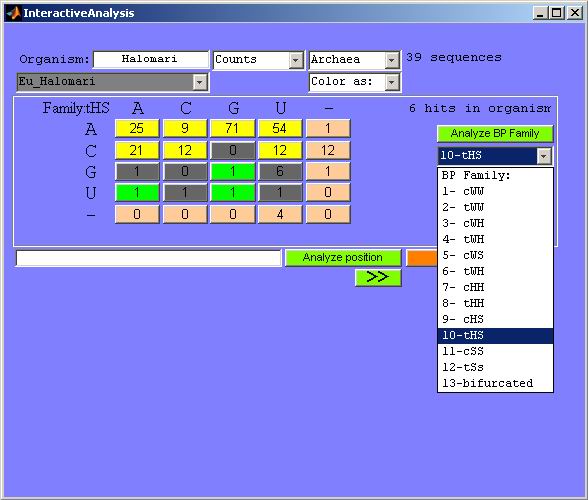
Figure 20. Analysis of all occurrences of a
basepair family. All basepairs of family 10 (tHS) are analyzed here in the
archaeal
5S rRNA alignment.
The GUI shown in Figure 20 states on the top right the number
of sequences in the chosen domain (in this case the domain archeae has 39 sequences).
Below that, the number of basepairs forming this particular interaction in
the source organism is shown. In this case six such tHS basepairs are found.
The isosterically colored buttons that appeared in the middle of the GUI display
the substitutions in the sequence positions corresponding to all these six
positions at once. Notice how most of them are clumped in the yellow isosteric
subfamily (the colors are the same as defined previously for the HTML output
files). Upon clicking on any of these colored buttons the names of sequences
giving rise to them (preceded by the corresponding nucleotide numbers in the
source organism) will be displayed in the lower part of the GUI (Figure 21).
This allows for easy identification of organisms with potential mistakes in
their alignments, so that the investigator can realign them by hand.
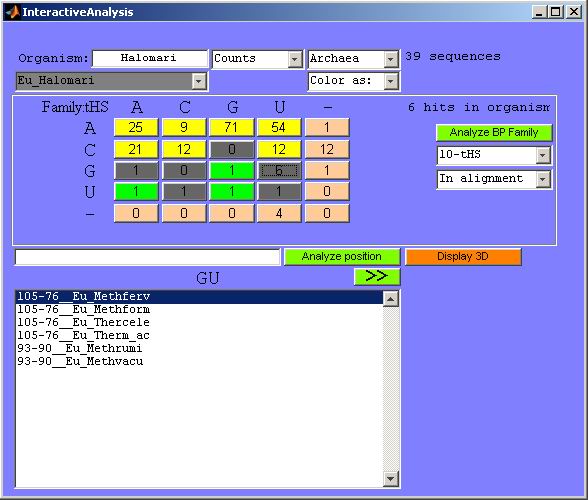
Figure 21. Getting sequence names with specific
substitution patterns. Upon clicking any button in the substitution matrix
(button GU
is clicked here), the names of sequences giving rise to the substitutions
become displayed in the lower part of the GUI. Sequence names are preceded
by the corresponding nucleotide numbers from the source organism.
4.2. Interactive analysis of individual basepairs or motifs
In addition to analyzing a family of basepairs, it is possible through this
GUI to analyze a particular position by entering individual numbers separated
by commas “,” or dashes “-” into the editable box in
the middle of the GUI. But first, the source of local nucleotide numbers has
to be specified. If this GUI is activated after a BP or NT list is loaded,
then the sequence name of the first entry will be initially displayed in the
white editable box and gray drop-menu box that define the source sequence for
the analysis (upper left corner of the GUI). Both of these boxes can be used
to specify the source organism, but in case the sequence names in these two
boxes are different, then the white editable box takes priority. If universal
numbers are desired the word “universal” should be typed in the
white editable box and carriage return pressed. If only two
nucleotide numbers separated by “,” are entered (such as “22,
26”), the sequence substitutions for these two positions are displayed
in the same output format seen for the basepair family analysis described in
the previous section. The value in the box that corresponds to the source organism
will be printed in bold. If the nucleotide numbers entered are present in the
input Excel BP list and the interaction they make is known, the buttons will
be colored according to their BP family isostericity matrix. The family name
also will be printed on the upper left corner. If any of the buttons is clicked
the names of sequences giving rise to the value in it are displayed in the
lower part of the GUI (Figure 22). (Note: it is possible to scroll between
the basepairs from the input BP list one by one by clicking the green buttons
labeled “<<” or “>>” that appear in the
middle of the GUI).

Figure 22. Basepair interactive analysis. Notice
that if the query is “22, 26” instead of “26, 22” the
same result is obtained, since this is a known BP present in the input BP list
and
it is always colored in the same way. The BP identity present in the source
organism (also the crystal structure) is CG (printed in bold font).
If instead of a basepair position (such as “22, 26”),
the user enters only one, or more that two numbers separated by commas, or
if the user enters two numbers separated by a dash, or any logical combination
of comma-separated and dash-separated numbers, then a motif sequence analysis
will be performed and results will be produced in a different format, where
the motif pattern and its number of occurrences will be printed in text instead
of the colored table. The pattern present in the source sequence will also
be indicated by the “<<<” sign. Once again, if the user
clicks on any one of the nucleotide patterns, the names of sequences giving
rise to them will be displayed in the lower portion of the GUI (Figure 23).

Figure 23. Motif interactive analysis. Individual
nucleotides in the motif are arranged in the order of the positions entered.
Since the query here is “26-22, 37”, the letters that correspond
to local number 26 of source organism is displayed first, then those that
correspond for 25, then 24, 23, 22, and finally 37. If instead of this the
query was “22-26, 37” the result would be displayed differently.
Finally, the interactive GUI provides a quick link to a basic
structure viewer through the activation of the button “Display 3D”.
When this button is clicked the first time it prompts the user to choose the
PDB file corresponding to the sequence studied, and atomic coordinates are
read (the bigger the PDB file the slower this process). The structure of the
input nucleotides is then shown. Subsequent activations of the button within
the same session display the structure immediately.
4.3.
Notes
1- The user can change the color distribution of the substitution
table to be similar to another basepair family. This may be important in
case mistakes in the BP list are suspected, or in case no BP family is stated,
causing the whole table to be in one neutral color.
2- It has been shown that the colored GUI buttons may display the results
of either a whole BP family or an individual position. To prevent any confusion,
a thin white box appears everytime one of these two types of analyses is
requested. This line surrounds the output buttons as well as the input buttons
giving rise to them. The difference can be clearly seen between Figures 20
and 21 on the one hand and Figures 22 and 23 on the other.
5. Ribostral
preferences
Ribostral is a diverse program capable of dealing with data in a variety
of ways. Through its preferences, the user can enhance his experience with
the program and make it do what is best for the investigation in hands. Ribostral
preferences can be accessed from the File menu of the main GUI. They are
then saved as a data file on the users disk, for easy passing between all
subordinate Ribostral GUIs. Last positions of GUIs before exiting them are
also saved in this file, so that GUIs always open in the last known position
they were closed in. Figure 24 shows the default preferences of the program.
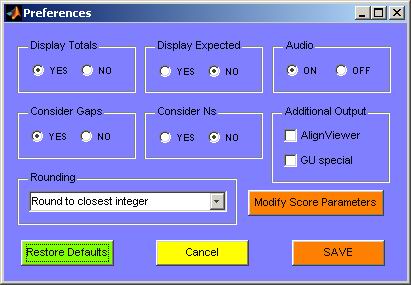
Figure 24. Ribostral default preferences.
The preferences are divided into several categories. Round
radio buttons belonging to the same category are mutually exclusive. The first
two categories, “Display Totals” and “Display Expected”,
affect only the list output files and do not affect the interactive analysis
GUI. If “Display Totals” is enabled, the totals of each row and
column of the BP list output are printed with the output (as in Figure 13).
Otherwise, the totals will be left out. If “Display Expected” is
enabled, the expected value for each substitution is printed in parentheses
after the observed substitution value, as in Figure 25.
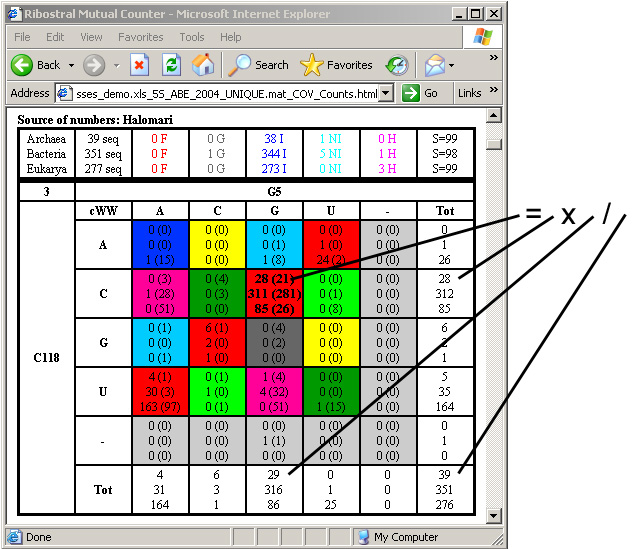
Figure 25. Substitution counts with expected
values (in parentheses).
The expected value for each box in the table is obtained by applying the formula:
Expected (of box in row a, column b) = Sum of boxes in row a * Sum of boxes
in columns b / Table total
This value is a measure of the likelihood of observing substitutions in the
specific box, based on the values observed in the neighboring boxes.
Enabling the “Audio” option allows the program to play specific
musical chimes every time an operation is completed or an error occurs. This
is useful in case a large file is being read or analyzed. In such cases the
user will be notified by the sound to carry on to the next step.
Enabling “Consider Gaps” means that the program will not ignore
insertion characters (“–”) seen in the alignment. Disabling
this option not only stops the program from showing gaps in the output, it
also ignores gaps from any other calculations, such as score or percent substitution
calculations. The “consider Ns” option does the same thing but
with characters representing undetermined nucleotides (usually symbolized by “o”, “O”, “n”,
and “N”, which as stated earlier are all converted to “o” when
the FASTA alignment file is originally read). These two options do not affect
analysis of non-basepairs, such as base triples or longer motifs.
Several rounding options are available through a dropdown menu. The choice
here affects the presentation of all decimal calculations done by Ribostral.
Activating any of the “Additional Output” options results in the
production of one or two more output formats that Ribostral does not produce
by default (discussed earlier). Preferences are saved only if the “SAVE” button
is activated.
Finally, upon activating the button “Modify Score Parameters”,
the file containing score parameters (“<installation directory>\Ribostral\SCORES.txt”)
is opened for editing. The parameters entered in this file affect all score
calculations. The resulting formula is shown on screen and is also printed
out in some output files every time sequence analysis is carried out.
6. Supporting tools
Ribostral provides an expandable sequence and structure analysis platform.
Additional tools can be easily and smoothly integrated into the main program.
There are currently five additional tools that allow Ribostral to do operations
beyond structurally analyzing sequence alignments (Figure 26). Each of these
tools is discussed separately below.
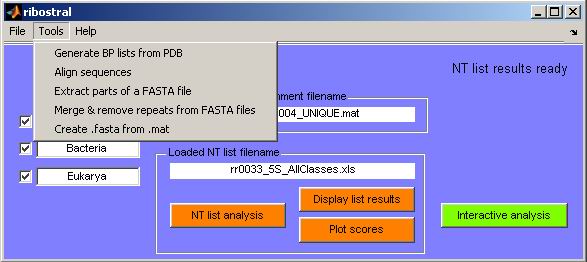
Figure 26. Ribostral tools.
6.1.
Tool 1: Generate BP list from PDB
This tool currently works only on PC platforms. PDB files are text files containing
all the atomic coordinates and related information about a structure. By analyzing
this data, interacting bases can be identified and classified into geometric
families (7). Upon activation of this tool, the user first chooses a PDB file
(browsing starts from the default PDB directory associated with Ribostral “<installation
directory>\PDB_structures”). Then, the user is prompted to enter the
sequence name in the alignment that corresponds to this 3D structure. Ribostral
then creates an Excel BP list for each of the basepair families found in the
crystal structure. It also creates a separate Excel BP list containing all
of them, and another Excel BP list containing all the non-cWW ones among them
together. These lists are created in the proper format to be read directly
by Ribostral and are saved in the default “<installation directory>\NT_lists” folder.
Note that the automatic classification of basepairs is not 100% accurate, so
the resulting automatic lists should be visually compared to the 3D structure
and corrected where needed.
6.2. Tool 2: Align sequences
This tool opens a GUI that interfaces with automatic motif alignment programs
based on a hybrid Stochastic Context Free Grammars/Markov Random Field (SCFG/MRF)
model. The GUI facilitates some of the steps in running these programs, and
allows the user to parse and align motifs that have their SCFG/MRF nodes
already characterized based on known 3D structure. Currently, MATLAB script
files still
need to be modified in order to parse and align new motifs. This tool mainly
provides an easily expandable user-friendly platform for applying automatic
sequence alignment.
6.3. Tool
3: Extract parts of a FASTA file
This tool, shown in Figure 27, requires loading a FASTA file from Ribostral
main GUI before it can be activated.
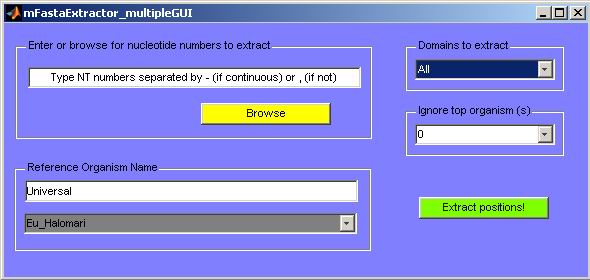
Figure 27. GUI for extracting parts of a FASTA
file.
The user then simply needs to indicate the motif of interest by entering its
nucleotide numbers in the editable box. All positions homologous to these numbers
will be extracted from the original alignment and saved as a new sub-alignment
in FASTA format. If there are one or more commas separating two or more groups
of numbers, the sub-alignment will represent these breaks in continuity by
four consecutive dots (....). This continuity separation code is understood
by other Ribostral tools, such as the alignment parser tool discussed in the
previous section. Naturally, nucleotide numbers refer to the source or reference
organism specified in the GUI. Similarly to the interactive analysis GUI, in
case there is disagreement between the sequence name in the white editable
box and that in the gray drop-menu box, the white editable box takes precedence.
The user can choose to ignore up to five organisms from the top of the sequence
alignment (for instance, if these are structure masks or sequences that do
not belong to the same phylogenetic group), and can choose to extract one domain
or all domains at once. The extracted sub-alignment is saved in the same directory
as the original source alignment file.
If there is more than one motif to be extracted, the user
can create a tab-delimited input text file containing the names of all of them
and their nucleotide
numbers for batch processing. The nucleotide numbers should correspond
to the Reference Organism Name selected in the GUI. The input text file would
look like this:
| h5IL |
|
53-56,356-358 |
| h6IL |
|
64-69,99-103 |
| h7IL1 |
|
128-131,231-233 |
| h7IL2 |
|
133-136,227-229 |
The nucleotide positions are then extracted
from the alignment the same way as if they were entered one by one in the GUI
editable
box. An
advantage
here is that the user can assign specific names to the sub-alignments containing
these motifs, instead of the otherwise generic names derived from nucleotide
numbers extracted.
6.4.
Tool 4: Merge & remove repeats from FASTA files
This tool opens a new GUI from which the user can browse for a minimum of one
and a maximum of three input FASTA files. Each one of the files read will be
checked for individual sequence lengths and repeats in sequence names. If some
sequence lengths are shorter than others, gap characters (“–”)
are added to the ends of the short ones. This prevents some errors when the
alignments are analyzed. If name repeats are found (the name meant here is
the first separate word after the “>” sign in a FASTA header
line), only the sequence with most valid nucleotide letters (A, C, G, and U)
is kept (it is possible to change this default criteria and choose sequences
with shortest internal gaps; this can be done by changing the value for the
variable “Criteria” in the script file mFastaPrinter.m). The resulting “unique” alignment
represents a more divergent set of phylogenetic taxa which may be more meaningful
to analyze. The “unique” FASTA files are saved in the same directory
and under the same names (with descriptive suffixes) as the original FASTA
files they are created from. If more than one original file was inputted into
this tool, a merged file containing all of them is also created.
6.5. Tool 5: Create
.fasta from .mat
This is a simple tool that allows the user to open a “*.mat” file
representing a sequence alignment in MATLAB data format and recreate the “*.fasta” file
it was originally created from. If the file contains more than one subgroup
(or phylogenetic domain), separate FASTA files containing the sequence of each
of them, in addition to one containing them all together, are created and saved
in the same directory as the input “*.mat” file.
7. Help menu
Under the help menu, the manual, website, and version information of Ribostral
can be found. To check if any updated version of the program is available,
the user can compare the local version number to the version number listed
on Ribostral website.
8. Sample study
In this section, a simple example of how Ribostral can be used to improve a
sequence alignment based on structure is given. We will do this exercise on
a small part of an alignment to keep things simple and clear. This is the starting
alignment:
>Structure
CGCCA-....-UAAGG
>Seq2
CGCCU-....-AAAUG
>Seq3
GGCAU-....-AAGGC
>Seq4
GGCAC-....-GGGUC
>Seq5
CACAC-....-GGGUG
>Seq6
CGCAG-....-CAGUG
>Seq7
AGUCG-....-CGCUG
>Seq8
UACAC-....-GGAUA
>Seq9
UGUCA-....-UAUGA
This is an alignment of an internal loop motif similar to
one seen in 16S rRNA internal loop 20 (IL20), the four small dots in the middle
are just to indicate strand dicontinuity (Ribostral
will consider points like this in an alignment as N's, in other words, they
are assigned local numbers unlike the gap symbol "-").
It is hard to tell whether this is a good alignment or not by just looking
at
it.
Some
purines
and pyrimidines
"seem" to be aligned well, there also
seems
to be a couple of cWW interactions that are covarying nicely, but
it is difficult to tell anything specific. Suppose that we know the 3D structure
of the
first sequence seen in this alignment (the one labeled "Structure"),
and that
it looks as in Figure 28:

Figure 28. Structure representation of the
first sequence in the alignment. Althought the sequence alignment is modified
to simplify the example, this motif does occur several times in rRNA, as in
internal loop 20 (IL20) of the 16S rRNA.
This structure can easily be encoded in Excel in
the form that Ribostral can handle. Figure 29 shows how this file looks like:

Figure 29. The same information in Figure 28
(BPs observed in 3D structure) written into Excel in the form required by Ribostral.
Given this structural information, we can objectively
measure the quality of the alignment. All that
needs
to be done
is to load
both
the FASTA
file and the Excel file into Ribostral, and to
analyze the output files produced. We will first look at the HTML Alignment
Viewer file,
which
displays
the alignment
with nucleotides colored in a way describing how well they conform to isostericity
rules inferred from structure:
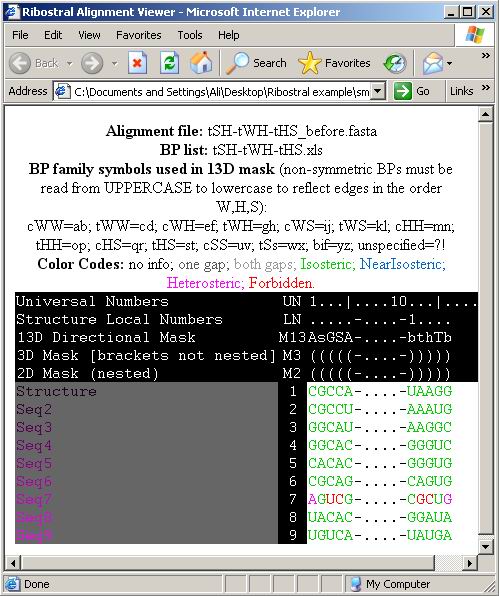
Figure 30. The HTML Alignment Viewer output,
showing areas
where the alignment is not consistent with structure (red and pink areas of Seq7).
So clearly, this alignment is not perfect, and it needs some
enhancement based on what is isosterically acceptable in 3D and what is not
(the
score of this alignment based on the formula described in section 3.2
above is 87).
The sequence
that is the cause of the problem is also clearly identified (Seq7). To fix
the alignment, we need to edit it in a sequence editor or text editor where
nucleotides can be moved by
hand (an automatic aligner based on Stochastic Context Free Grammar/Markov
Random Fields or SCFG/MRF is in preparation; Mokdad, A., Sarver, M., Stombaugh,
J., Zirbel, C., Leontis, N.B.). Ribostral also provides some insight on how
the alignment
should be fixed through its "_Cov.html" output files (where a substitution
table for each BP position is provided with isosterically inspired colored).
In this
particular case, and because of the specific BP interactions formed in
this motif, a small manipulation of Seq7 second part (after the dots) creates
an insertion, but fixes problems with forbidden and heterosteric BPs.
After this manipulation, the resulting alignment was viewed again with the
HTML Alignment Viewer, and Figure 31 shows the results:
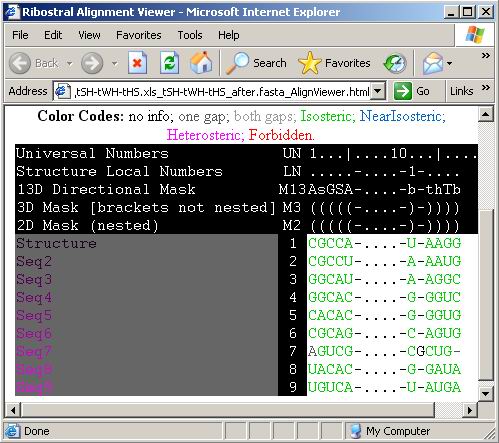
Figure 31. The corrected alignment as seen
again with the Alignment Viewer.
Obviously, Figure 31 shows an improvement in the alignment
(new score = 96), eventhough it created an insertion in Seq7. The manipulation
also created an addition gap on the far right of Seq7, but this may not be
a true gap because depending on
the unseen
nucleotides
that
are present beyond this motif (remember, for simplicity the alignment in this
example is just an small extract from a bigger alignment). The reason the
alignment was enhanced to this degree is the fact that the CUG- at
the end of Seq7 fit better the isostericity rules of the tSH, tWH, tHS, and
cWW BPs they now occupy
(respectively),
as compared to the GCUG.
References:
1. Hall, T.A. (1999). BioEdit: a user-friendly biological sequence alignment
editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser.,
41, 95-98.
2. Leontis, N.B. and Westhof, E. (2001). Geometric nomenclature and classification
of RNA base pairs. RNA, 7, 499-512.
3. Jossinet, F. and Westhof, E. (2005). Sequence to Structure (S2S): display,
manipulate and interconnect RNA data from sequence to structure. Bioinformatics,
21, 3320-3321.
4. Leontis, N.B., Stombaugh, J. and Westhof, E. (2002). The non-Watson-Crick
base pairs and their associated isostericity matrices. Nucleic Acids Res, 30,
3497-3531.
5. Lemieux, S. and Major, F. (2002). RNA canonical and non-canonical base pairing
types: a recognition method and complete repertoire. Nucleic Acids Res, 30,
4250-4263.
6. Mokdad, A., Krasovska, M.V., Sponer, J. and Leontis, N.B. (2006). Structural
and evolutionary classification of G/U wobble basepairs in the ribosome. Nucleic
Acids Res, 34, 1326-1341.
7. Sarver M., Zirbel C., Stombaugh J., Mokdad A., Leontis N. (2006). Finding
Local and Composite Recurrent Structural Motifs in RNA 3D Structure. Journal
of Mathematical Biology (accepted into special RNA issue).
Citation:
Please cite the following work if you use
any component of Ribostral or the nomenclatures proposed in it:
Mokdad, A., and Leontis, N. (2006). Ribostral: An RNA 3D alignment analyzer
and viewer based on basepair isostericities. Bioinformatics, 22(17): 2168-70.

















 Figure
17. Ribostral HTML alignment viewer. The visible part of the 5S alignment
describes how well sequences agree with structure, represented here by Eu_Halomari,
or sequence number 11. The color code is printed at the top. Note that the
whole content including colors can be copy/pasted into Excel or other editors
for manipulation.
Figure
17. Ribostral HTML alignment viewer. The visible part of the 5S alignment
describes how well sequences agree with structure, represented here by Eu_Halomari,
or sequence number 11. The color code is printed at the top. Note that the
whole content including colors can be copy/pasted into Excel or other editors
for manipulation.












