Motif IL_17948.1 Version IL_17948.1 of this group appears in releases 3.91 to 3.94
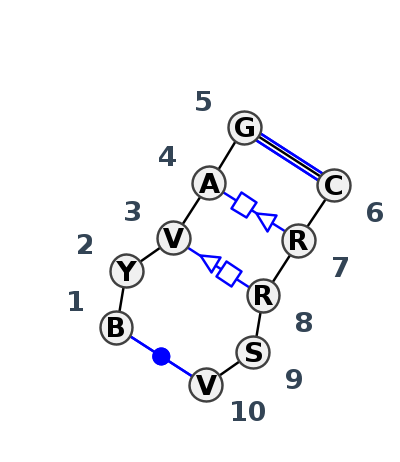
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 10 | 1-10 | 2-9 | 3-8 | 4-7 | 5-6 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_5J7L_056 | 5J7L | 0.3806 | 0 | Double sheared with non-canonical cWW | AA | SSU rRNA | G | 1272 | C | 1273 | A | 1274 | A | 1275 | G | 1276 | * | C | 1259 | G | 1260 | A | 1261 | C | 1262 | C | 1263 | cWW | ncWB | tSH | tHS | cWW |
| 2 | IL_4V9F_085 | 4V9F | 0.2352 | 0 | Double sheared with non-canonical cWW | 0 | LSU rRNA | G | 2501 | C | 2502 | A | 2503 | A | 2504 | G | 2505 | * | C | 2515 | G | 2516 | A | 2517 | C | 2518 | C | 2519 | cWW | cWB | ntSH | tHS | cWW |
| 3 | IL_4WF9_093 | 4WF9 | 0.1248 | 0 | Double sheared with non-canonical cWW | X | LSU rRNA | C | 2493 | C | 2494 | A | 2495 | A | 2496 | G | 2497 | * | C | 2507 | G | 2508 | A | 2509 | C | 2510 | G | 2511 | cWW | cWB | tSH | ntHS | cWW |
| 4 | IL_5J7L_338 | 5J7L | 0.0000 | 0 | Double sheared with non-canonical cWW | DA | LSU rRNA | C | 2466 | C | 2467 | A | 2468 | A | 2469 | G | 2470 | * | C | 2480 | G | 2481 | A | 2482 | C | 2483 | G | 2484 | cWW | cWB | ntSH | tHS | cWW |
| 5 | IL_8P9A_333 | 8P9A | 0.1090 | 0 | Double sheared with non-canonical cWW | AR | LSU rRNA | U | 2835 | C | 2836 | A | 2837 | A | 2838 | G | 2839 | * | C | 2849 | G | 2850 | A | 2851 | C | 2852 | A | 2853 | cWW | tSH | tHS | cWW | |
| 6 | IL_7A0S_087 | 7A0S | 0.1399 | 0 | Double sheared with non-canonical cWW | X | LSU rRNA | C | 2445 | C | 2446 | G | 2447 | A | 2448 | G | 2449 | * | C | 2459 | G | 2460 | G | 2461 | C | 2462 | G | 2463 | cWW | ncWB | tHS | cWW | |
| 7 | IL_9DFE_095 | 9DFE | 0.2144 | 0 | Double sheared with non-canonical cWW | 1A | LSU rRNA | C | 2466 | C | 2467 | G | 2468 | A | 2469 | G | 2470 | * | C | 2480 | G | 2481 | G | 2482 | C | 2483 | G | 2484 | cWW | cWB | tHS | cWW | |
| 8 | IL_4WFL_001 | 4WFL | 0.2160 | 0 | Double sheared with non-canonical cWW | A | LSRP RNA | U | 72 | C | 73 | G | 74 | A | 75 | G | 76 | * | C | 91 | G | 92 | A | 93 | C | 94 | A | 95 | cWW | cWB | tSH | tHS | cWW |
| 9 | IL_5J7L_274 | 5J7L | 0.2585 | 0 | Double sheared with non-canonical cWW | DA | LSU rRNA | G | 875 | C | 876 | A | 877 | A | 878 | G | 879 | * | C | 898 | A | 899 | A | 900 | C | 901 | C | 902 | cWW | cWB | ntSH | tHS | cWW |
| 10 | IL_7A0S_056 | 7A0S | 0.4089 | 0 | Double sheared with near cWW | X | LSU rRNA | C | 1529 | U | 1530 | C | 1531 | A | 1532 | G | 1533 | * | C | 1491 | A | 1492 | A | 1493 | G | 1494 | G | 1495 | cWW | tHS | ncWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GCAAG*CGACC | 2 |
| CCAAG*CGACG | 2 |
| CCGAG*CGGCG | 2 |
| UCAAG*CGACA | 1 |
| UCGAG*CGACA | 1 |
| GCAAG*CAACC | 1 |
| CUCAG*CAAGG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CAA*GAC | 5 |
| CGA*GGC | 2 |
| CGA*GAC | 1 |
| CAA*AAC | 1 |
| UCA*AAG | 1 |
- Annotations
-
- Double sheared with non-canonical cWW (9)
- Double sheared with near cWW (1)
- Basepair signature
- cWW-L-R-tSH-tHS-cWW
- Heat map statistics
- Min 0.11 | Avg 0.26 | Max 0.51
Coloring options: