Motif HL_68304.1 Version HL_68304.1 of this group appears in releases 3.2 to 3.2
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 1-6 | 2-5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_3KTW_001 | 3KTW | 0.3622 | 0 | GNRA | C | SRP RNA | U | 163 | G | 164 | G | 165 | A | 166 | G | 167 | G | 168 | cWW | |
| 2 | HL_4WF9_031 | 4WF9 | 0.0000 | 0 | GNRA | X | LSU rRNA | C | 1260 | G | 1261 | U | 1262 | A | 1263 | A | 1264 | G | 1265 | cWW | |
| 3 | HL_4IOA_030 | 4IOA | 0.2218 | 0 | GNRA | X | LSU rRNA | C | 1235 | G | 1236 | G | 1237 | A | 1238 | A | 1239 | G | 1240 | cWW | |
| 4 | HL_4LFB_012 | 4LFB | 0.7503 | 1 | GNRA | A | SSU rRNA | C | 458 | G | 459 | A | 460 | G | 462 | A | 463 | G | 474 | cWW | tSH |
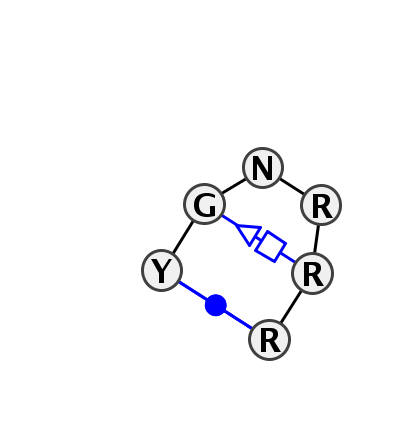
| 5 | HL_5J7L_148 | 5J7L | 0.7835 | 1 | GNRA | DA | LSU rRNA | C | 487 | G | 488 | G | 489 | G | 491 | A | 492 | G | 493 | cWW | tSH |
| 6 | HL_5J7L_026 | 5J7L | 0.8285 | 1 | GNRA | AA | SSU rRNA | U | 1165 | G | 1166 | A | 1167 | A | 1169 | A | 1170 | A | 1171 | cWW | tSH |
| 7 | HL_5J7L_180 | 5J7L | 0.8060 | 0 | GNRA variation | DA | LSU rRNA | C | 1868 | G | 1869 | C | 1870 | A | 1871 | A | 1872 | G | 1873 | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UGGAGG | 1 |
| CGUAAG | 1 |
| CGGAAG | 1 |
| CGACGAG | 1 |
| CGGCGAG | 1 |
| UGAUAAA | 1 |
| CGCAAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GGAG | 1 |
| GUAA | 1 |
| GGAA | 1 |
| GACGA | 1 |
| GGCGA | 1 |
| GAUAA | 1 |
| GCAA | 1 |
Release history
| Release | 3.2 |
|---|---|
| Date | 2018-09-26 |
| Status | New id, no parents |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- GNRA (6)
- GNRA variation (1)
- Basepair signature
- cWW-tSH-R-R
- Heat map statistics
- Min 0.22 | Avg 0.60 | Max 0.96
Coloring options: