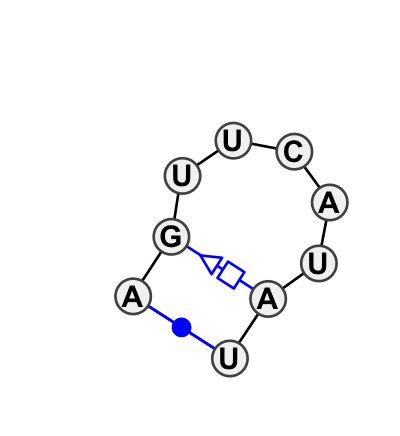
Motif HL_96039.1 Version HL_96039.1 of this group appears in releases 0.1 to 0.2
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 1-9 | 2-8 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_1S72_062 | 1S72 | 0.3320 | 0 | 0 | LSU rRNA | A | 2506 | G | 2507 | C | 2508 | A | 2509 | C | 2510 | A | 2511 | U | 2512 | A | 2513 | U | 2514 | cWW | tSH |
| 2 | HL_2QBG_062 | 2QBG | 0.0000 | 0 | B | LSU rRNA | A | 2471 | G | 2472 | U | 2473 | U | 2474 | C | 2475 | A | 2476 | U | 2477 | A | 2478 | U | 2479 | cWW | tSH |
| 3 | HL_3O58_062 | 3O58 | 0.2679 | 0 | 1 | LSU rRNA | C | 2840 | G | 2841 | U | 2842 | U | 2843 | C | 2844 | A | 2845 | U | 2846 | A | 2847 | G | 2848 | cWW | ntSH |
| 4 | HL_3PYO_060 | 3PYO | 0.2681 | 0 | A | LSU rRNA | C | 2471 | G | 2472 | U | 2473 | C | 2474 | C | 2475 | A | 2476 | C | 2477 | A | 2478 | G | 2479 | cWW | tSH |
| 5 | HL_2ZJR_058 | 2ZJR | 0.3877 | 0 | X | LSU rRNA | A | 2450 | G | 2451 | U | 2452 | C | 2453 | C | 2454 | A | 2455 | U | 2456 | A | 2457 | U | 2458 | cWW | tSH |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AGCACAUAU | 1 |
| AGUUCAUAU | 1 |
| CGUUCAUAG | 1 |
| CGUCCACAG | 1 |
| AGUCCAUAU | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GUUCAUA | 2 |
| GCACAUA | 1 |
| GUCCACA | 1 |
| GUCCAUA | 1 |
Release history
Parent motifs
This motif has no parent motifs.
Children motifs
| Child motif | Common motif instances | Only in HL_96039.1 | Only in the child motif |
|---|---|---|---|
| HL_46489.1 Compare | HL_1S72_062, HL_2ZJR_058, HL_2QBG_062 | HL_3PYO_060, HL_3O58_062 | HL_4A1B_061, HL_3V2F_058, HL_3U5H_062 |
Coloring options: