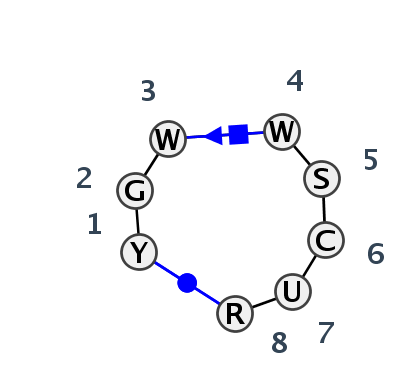
Motif HL_99167.2 Version HL_99167.2 of this group appears in releases 3.84 to 3.86
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 1-8 | 3-4 | 3-5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_2XD0_006 | 2XD0 | 0.4830 | 2 | Pseudoknot with intercalation | W | TOXI | U | 4 | G | 5 | U | 8 | U | 9 | G | 10 | C | 11 | U | 12 | A | 13 | cWW | ncSH | |
| 2 | HL_7D8O_001 | 7D8O | 0.4593 | 2 | Pseudoknot with intercalation | B | Antitoxin RNA | U | 4 | G | 5 | U | 8 | U | 9 | G | 10 | C | 11 | U | 12 | A | 13 | cWW | ncSH | |
| 3 | HL_2XDB_001 | 2XDB | 0.4624 | 2 | Pseudoknot with intercalation | G | TOXI | U | 4 | G | 5 | U | 8 | U | 9 | G | 10 | C | 11 | U | 12 | A | 13 | cWW | ncSH | |
| 4 | HL_7A0S_001 | 7A0S | 0.1996 | 2 | Pseudoknot with intercalation | X | LSU rRNA | C | 58 | G | 59 | U | 62 | A | 63 | C | 64 | C | 65 | U | 66 | G | 67 | cWW | cSH | |
| 5 | HL_4V9F_001 | 4V9F | 0.1533 | 2 | Pseudoknot with intercalation | 0 | LSU rRNA | U | 55 | G | 56 | A | 59 | A | 60 | G | 61 | C | 62 | U | 63 | G | 64 | cWW | cSH | |
| 6 | HL_7RQB_001 | 7RQB | 0.0000 | 2 | Pseudoknot with intercalation (H) | 1A | LSU rRNA | U | 59 | G | 60 | U | 63 | A | 64 | C | 65 | C | 66 | U | 67 | G | 68 | cWW | ncSH |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UGAUUUGCUA | 3 |
| CGAUUACCUG | 1 |
| UGCCAAGCUG | 1 |
| UGGCUACCUG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAUUUGCU | 3 |
| GAUUACCU | 1 |
| GCCAAGCU | 1 |
| GGCUACCU | 1 |
- Annotations
-
- Pseudoknot with intercalation (5)
- Pseudoknot with intercalation (H) (1)
- Basepair signature
- cWW-F-F-cSH-F-F
- Heat map statistics
- Min 0.07 | Avg 0.27 | Max 0.48
Coloring options: