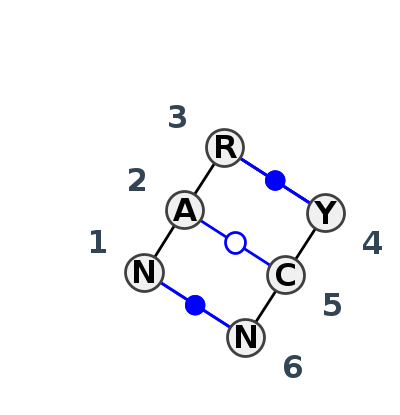
Motif IL_10432.1 Version IL_10432.1 of this group appears in releases 4.0 to 4.0
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | break | 4 | 5 | 6 | 1-6 | 2-5 | 3-4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_4V9F_060 | 4V9F | 0.0993 | 0 | Isolated tWW turn | 0 | LSU rRNA | U | 1741 | A | 1742 | G | 1743 | * | C | 2036 | C | 2037 | A | 2038 | cWW | tWW | cWW |
| 2 | IL_9E6Q_062 | 9E6Q | 0.0743 | 0 | Isolated tWW turn (H) | 1 | LSU rRNA | C | 1807 | A | 1808 | G | 1809 | * | OMC | 2116 | C | 2117 | G | 2118 | cWW | tWW | cWW |
| 3 | IL_8P9A_303 | 8P9A | 0.0000 | 0 | AR | LSU rRNA | A | 1895 | A | 1896 | G | 1897 | * | C | 2338 | C | 2339 | U | 2340 | cWW | tWW | cWW | |
| 4 | IL_8OI5_077 | 8OI5 | 0.0642 | 0 | 1 | LSU rRNA | A | 1891 | A | 1892 | G | 1893 | * | C | 2316 | C | 2317 | U | 2318 | cWW | tWW | cWW | |
| 5 | IL_9AXU_071 | 9AXU | 0.0890 | 0 | 2 | LSU rRNA | A | 1950 | A | 1951 | G | 1952 | * | C | 2426 | C | 2427 | U | 2428 | cWW | tWW | cWW | |
| 6 | IL_4WF9_063 | 4WF9 | 0.0965 | 0 | Isolated tWW turn | X | LSU rRNA | U | 1707 | A | 1708 | A | 1709 | * | U | 2022 | C | 2023 | A | 2024 | cWW | tWW | cWW |
| 7 | IL_9DFE_064 | 9DFE | 0.1116 | 0 | 1A | LSU rRNA | C | 1663 | A | 1664 | A | 1665 | * | U | 1995 | C | 1996 | G | 1997 | cWW | tWW | cWW | |
| 8 | IL_7A0S_060 | 7A0S | 0.1032 | 0 | Isolated tWW turn | X | LSU rRNA | U | 1680 | A | 1681 | A | 1682 | * | U | 1978 | C | 1979 | A | 1980 | cWW | tWW | cWW |
| 9 | IL_8B0X_124 | 8B0X | 0.1256 | 0 | Isolated tWW turn (H) | a | LSU rRNA | G | 1665 | A | 1666 | A | 1667 | * | U | 1999 | C | 2000 | C | 2001 | cWW | tWW | cWW |
| 10 | IL_8GLP_303 | 8GLP | 0.3105 | 0 | L5 | LSU rRNA | A | 2844 | A | 2845 | G | 2846 | * | C | 3842 | C | 3843 | PSU | 3844 | cWW | tWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AAG*CCU | 3 |
| UAA*UCA | 2 |
| UAG*CCA | 1 |
| CAG*(OMC)CG | 1 |
| CAA*UCG | 1 |
| GAA*UCC | 1 |
| AAG*CC(PSU) | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| A*C | 10 |
Release history
| Release | 4.0 |
|---|---|
| Date | 2025-08-13 |
| Status | New id, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (5)
- Isolated tWW turn (3)
- Isolated tWW turn (H) (2)
- Basepair signature
- cWW-tWW-cWW
- Heat map statistics
- Min 0.06 | Avg 0.14 | Max 0.35
Coloring options: