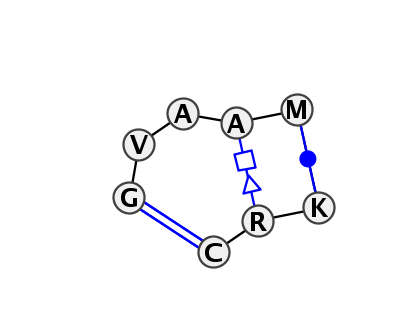
Motif IL_12566.1 Version IL_12566.1 of this group appears in releases 3.54 to 3.61
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 1-8 | 2-7 | 4-7 | 5-6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_4V9F_059 | 4V9F | 0.2364 | 2 | 0 | LSU rRNA | G | 1728 | A | 1729 | A | 1732 | A | 1733 | C | 1734 | * | G | 2045 | G | 2046 | C | 2047 | cWW | ncWW | tHS | cWW |
| 2 | IL_7RQB_064 | 7RQB | 0.0000 | 1 | 1A | LSU rRNA | G | 1651 | A | 1652 | A | 1654 | A | 1655 | C | 1656 | * | G | 2004 | A | 2005 | C | 2006 | cWW | tHS | cWW | |
| 3 | IL_7A0S_059 | 7A0S | 0.0799 | 1 | X | LSU rRNA | G | 1668 | A | 1669 | A | 1671 | A | 1672 | C | 1673 | * | G | 1987 | A | 1988 | C | 1989 | cWW | tHS | cWW | |
| 4 | IL_4WF9_062 | 4WF9 | 0.1426 | 1 | X | LSU rRNA | G | 1695 | C | 1696 | A | 1698 | A | 1699 | C | 1700 | * | G | 2031 | A | 2032 | C | 2033 | cWW | tHS | cWW | |
| 5 | IL_6D3P_003 | 6D3P | 0.9285 | 1 | A | RNA (45-MER) | G | 32 | G | 33 | A | 34 | A | 35 | A | 36 | * | U | 6 | A | 8 | C | 9 | cWW | tSH | cWW | |
| 6 | IL_6D3P_001 | 6D3P | 0.9285 | 1 | A | RNA (45-MER) | G | 32||||12_566 | G | 33||||12_566 | A | 34||||12_566 | A | 35||||12_566 | A | 36||||12_566 | * | U | 6||||12_566 | A | 8||||12_566 | C | 9||||12_566 | cWW | tSH | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GAGAAC*GAC | 2 |
| GGAAA*UAAC | 2 |
| GAGCAAC*GGC | 1 |
| GCGAAC*GAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AGAA*A | 2 |
| GAA*AA | 2 |
| AGCAA*G | 1 |
| CGAA*A | 1 |
Release history
| Release | 3.54 | 3.55 | 3.56 | 3.57 | 3.58 | 3.59 | 3.60 | 3.61 |
|---|---|---|---|---|---|---|---|---|
| Date | 2022-02-02 | 2022-03-02 | 2022-03-30 | 2022-04-27 | 2022-05-25 | 2022-06-22 | 2022-07-20 | 2022-08-17 |
| Status | New id, 2 parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (6)
- Basepair signature
- cWW-L-L-tHS-cWW
- Heat map statistics
- Min 0.08 | Avg 0.49 | Max 0.99
Coloring options: