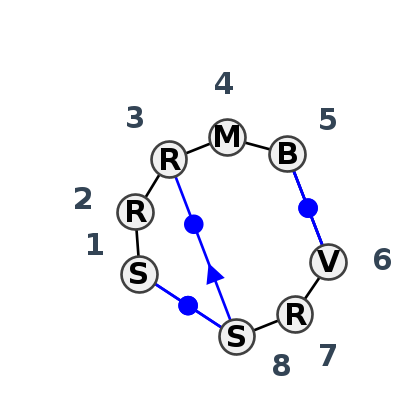
Motif IL_66997.1 Version IL_66997.1 of this group appears in releases 3.89 to 3.89
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 1-2 | 1-8 | 2-8 | 3-8 | 4-7 | 5-6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_4LFB_066 | 4LFB | 0.5223 | 1 | A | SSU rRNA | C | 1128 | A | 1130 | G | 1131 | C | 1132 | G | 1133 | * | C | 1141 | G | 1142 | G | 1143 | cWW | ncWS | ncWW | cWW | ||
| 2 | IL_5J7L_013 | 5J7L | 0.0000 | 0 | AA | SSU rRNA | G | 203 | G | 204 | A | 205 | C | 206 | C | 207 | * | G | 212 | G | 213 | C | 214 | cWW | cWW | ||||
| 3 | IL_4YAZ_005 | 4YAZ | 0.5243 | 1 | R | Cyclic di-GMP-I riboswitch | G | 77 | A | 78 | A | 79 | A | 80 | U | 81 | * | A | 4 | A | 6 | C | 7 | cSH | cWW | ntHS | cWS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CCAGCG*CGG | 1 |
| GGACC*GGC | 1 |
| GAAAU*ACAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CAGC*G | 1 |
| GAC*G | 1 |
| AAA*CA | 1 |
Release history
| Release | 3.89 |
|---|---|
| Date | 2024-10-09 |
| Status | New id, 2 parents |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (3)
- Basepair signature
- cWW-cWS-L-R-cWW-L
- Heat map statistics
- Min 0.52 | Avg 0.35 | Max 0.53
Coloring options: