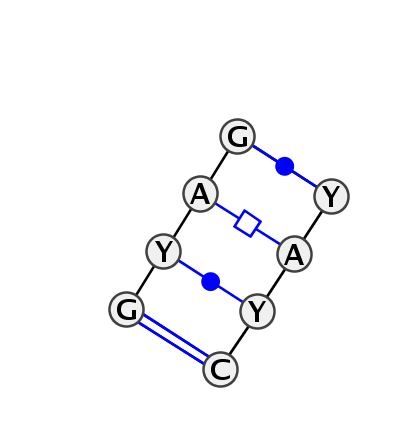
Motif IL_90423.1 Version IL_90423.1 of this group appears in releases 3.12 to 3.14
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | break | 5 | 6 | 7 | 8 | 1-8 | 2-7 | 3-6 | 4-5 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_3BNR_011 | 3BNR | 0.0000 | 1 | C+D | A site of human mitochondrial ribosome, chain one + A site of human mitochondrial ribosome, chain three | G | 15 | C | 16 | A | 17 | G | 19 | * | C | 5 | A | 6 | C | 7 | C | 8 | cWW | cWW | ||
| 2 | IL_5J7L_287 | 5J7L | 0.6444 | 1 | DA | LSU rRNA | G | 1202 | U | 1203 | A | 1204 | G | 1206 | * | U | 1240 | A | 1241 | U | 1242 | C | 1243 | cWW | cwW | tHH | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GCAAG*CACC | 1 |
| GUAAG*UAUC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CAA*AC | 1 |
| UAA*AU | 1 |
- Annotations
-
- (2)
- Basepair signature
- cWW-cWW-tHH-cWW
- Heat map statistics
- Min 0.64 | Avg 0.32 | Max 0.64
Coloring options: