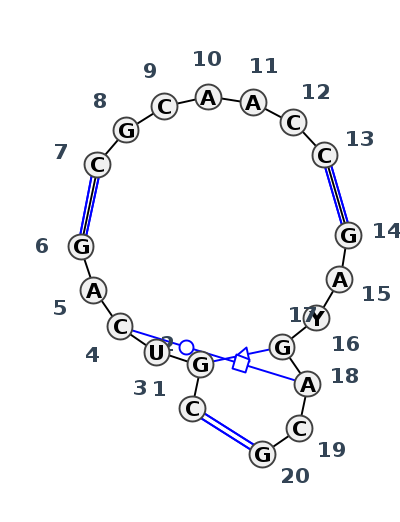
Motif J3_08394.3 Version J3_08394.3 of this group appears in releases 3.96 to 3.99
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | 10 | 11 | 12 | 13 | break | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 1-20 | 2-17 | 4-18 | 6-7 | 13-14 | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_4LFB_007 | 4LFB | 0.0000 | 0 | A | SSU rRNA | C | 1063 | G | 1064 | U | 1065 | C | 1066 | A | 1067 | G | 1068 | * | C | 1107 | G | 1108 | C | 1109 | A | 1110 | A | 1111 | C | 1112 | C | 1113 | * | G | 1187 | A | 1188 | C | 1189 | G | 1190 | A | 1191 | C | 1192 | G | 1193 | cWW | ntSS | tWH | cWW | cWW |
| 2 | J3_5J7L_008 | 5J7L | 0.1134 | 0 | AA | SSU rRNA | C | 1063 | G | 1064 | U | 1065 | C | 1066 | A | 1067 | G | 1068 | * | C | 1107 | G | 1108 | C | 1109 | A | 1110 | A | 1111 | C | 1112 | C | 1113 | * | G | 1187 | A | 1188 | U | 1189 | G | 1190 | A | 1191 | C | 1192 | G | 1193 | cWW | tSS | tWH | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGUCAG*CGCAACC*GACGACG | 1 |
| CGUCAG*CGCAACC*GAUGACG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GUCA*GCAAC*ACGAC | 1 |
| GUCA*GCAAC*AUGAC | 1 |
- Annotations
-
- (2)
- Basepair signature
- cWW-tSS-F-F-tWH-F-F-cWW-F-cWW-F-F-F-F-F
- Heat map statistics
- Min 0.11 | Avg 0.06 | Max 0.11
Coloring options: