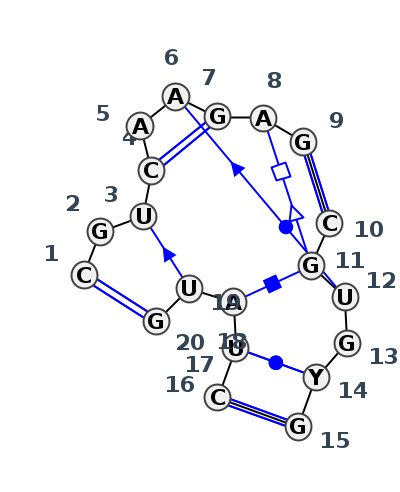
Motif J3_24131.1 Version J3_24131.1 of this group appears in releases 3.88 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | break | 10 | 11 | 12 | 13 | 14 | 15 | break | 16 | 17 | 18 | 19 | 20 | 1-20 | 3-19 | 4-7 | 4-18 | 6-12 | 8-11 | 9-10 | 11-18 | 14-17 | 15-16 | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_5TBW_060 | 5TBW | 0.0000 | 1 | 1 | LSU rRNA | C | 205 | G | 206 | U | 207 | C | 208 | A | 209 | A | 211 | G | 212 | A | 213 | G | 214 | * | C | 226 | G | 227 | U | 228 | G | 229 | U | 230 | G | 231 | * | C | 185 | U | 186 | A | 187 | U | 188 | G | 189 | cWW | csS | cWW | cSW | tHS | cWW | cHH | cWw | cWW | |
| 2 | J3_8C3A_046 | 8C3A | 0.1192 | 1 | 1 | LSU rRNA | C | 204 | G | 205 | U | 206 | C | 207 | A | 208 | A | 210 | G | 211 | A | 212 | G | 213 | * | C | 225 | G | 226 | U | 227 | G | 228 | C | 229 | G | 230 | * | C | 184 | U | 185 | A | 186 | U | 187 | G | 188 | cWW | csS | cWW | ncSs | cSW | tHS | cWW | cHH | ncWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CUAUG*CGUCAUAGAG*CGUGUG | 1 |
| CUAUG*CGUCACAGAG*CGUGCG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UAU*GUCAUAGA*GUGU | 1 |
| UAU*GUCACAGA*GUGC | 1 |
- Annotations
-
- (2)
- Basepair signature
- cWW-F-cSS-cHH-cWW-cWW-F-cWW-cSW-tHS-F-cWW
- Heat map statistics
- Min 0.12 | Avg 0.06 | Max 0.12
Coloring options: