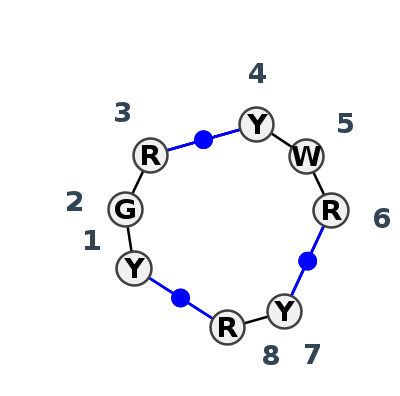
Motif J3_56052.3 Version J3_56052.3 of this group appears in releases 3.88 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | break | 4 | 5 | 6 | break | 7 | 8 | 1-8 | 3-4 | 6-7 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_4V88_080 | 4V88 | 0.6241 | 2 | A6 | SSU rRNA | U | 633 | G | 634 | A | 635 | * | U | 861 | A | 862 | A | 865 | * | U | 965 | A | 966 | cWW | cWW | cWW |
| 2 | J3_1G1X_001 | 1G1X | 0.2031 | 1 | D+E | 16S RIBOSOMAL RNA | C | 586 | G | 587 | G | 588 | * | C | 651 | U | 652 | G | 654 | * | C | 754 | G | 755 | cWW | cWW | cWW |
| 3 | J3_1KUQ_001 | 1KUQ | 0.1349 | 1 | B | 16S RIBOSOMAL RNA FRAGMENT | C | 4 | G | 5 | G | 6 | * | C | 15 | U | 16 | G | 18 | * | C | 53 | G | 54 | cWW | cWW | cWW |
| 4 | J3_5J7L_002 | 5J7L | 0.0000 | 1 | AA | SSU rRNA | C | 586 | G | 587 | G | 588 | * | C | 651 | U | 652 | G | 654 | * | C | 754 | G | 755 | cWW | cWW | cWW |
| 5 | J3_4LFB_011 | 4LFB | 0.1217 | 1 | A | SSU rRNA | C | 586 | G | 587 | G | 588 | * | C | 651 | U | 652 | G | 654 | * | C | 754 | G | 755 | cWW | cWW | cWW |
| 6 | J3_6CZR_016 | 6CZR | 0.2012 | 1 | 1a | SSU rRNA | C | 570 | G | 571 | G | 572 | * | C | 635 | U | 636 | G | 638 | * | C | 738 | G | 739 | cWW | ncWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGG*CUAG*CG | 4 |
| UGA*UAAUA*UA | 1 |
| CGG*CUUG*CG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| G*UA* | 4 |
| G*AAU* | 1 |
| G*UU* | 1 |
- Annotations
-
- (6)
- Basepair signature
- cWW-F-cWW-cWW-F
- Heat map statistics
- Min 0.12 | Avg 0.30 | Max 0.71
Coloring options: