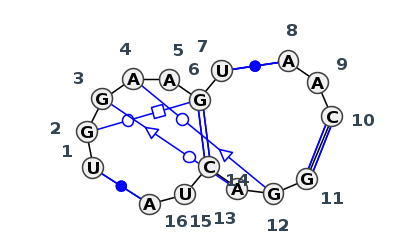
Motif J3_80847.1 Version J3_80847.1 of this group appears in releases 3.91 to 3.94
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | 10 | break | 11 | 12 | 13 | 14 | 15 | 16 | 1-16 | 2-6 | 3-13 | 4-12 | 6-14 | 7-8 | 10-11 | 15-16 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_8C3A_086 | 8C3A | 0.0000 | 2 | CM | SSU rRNA | U | 1393 | G | 1394 | G | 1395 | A | 1396 | A | 1397 | G | 1398 | U | 1401 | * | A | 1321 | A | 1322 | C | 1323 | * | G | 1372 | G | 1373 | A | 1374 | C | 1375 | U | 1376 | A | 1377 | cWW | tWH | tSW | tWS | cWW | cWW | cWW | ncSH |
| 2 | J3_8P9A_083 | 8P9A | 0.2099 | 2 | sR | SSU rRNA | U | 1407 | G | 1408 | G | 1409 | A | 1410 | A | 1411 | G | 1412 | U | 1415 | * | A | 1336 | A | 1337 | C | 1338 | * | G | 1386 | G | 1387 | A | 1388 | C | 1389 | U | 1390 | A | 1391 | cWW | tWH | tSW | tWS | cWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AAC*GGACUA*UGGAAGUUU | 2 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| A*GACU*GGAAGUU | 2 |
- Annotations
-
- (2)
- Basepair signature
- cWW-tWH-F-tSW-cWW-tWS-F-cWW-cWW-F
- Heat map statistics
- Min 0.21 | Avg 0.10 | Max 0.21
Coloring options: