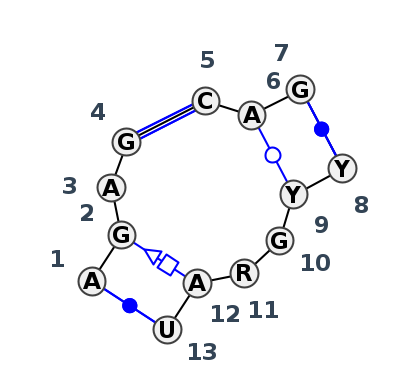
Motif J3_97642.2 Version J3_97642.2 of this group appears in releases 4.0 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | break | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 13 | 1-13 | 2-12 | 4-5 | 5-10 | 6-9 | 7-8 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_8GLP_013 | 8GLP | 0.1894 | 0 | S2 | SSU rRNA | A | 816 | G | 817 | A | 818 | G | 819 | * | C | 829 | A | 830 | G | 831 | * | C | 843 | U | 844 | G | 845 | G | 846 | A | 847 | U | 848 | cWW | tSH | cWW | cWW | ||
| 2 | J3_8P9A_073 | 8P9A | 0.0000 | 0 | sR | SSU rRNA | A | 760 | G | 761 | A | 762 | G | 763 | * | C | 773 | A | 774 | G | 775 | * | U | 785 | C | 786 | G | 787 | A | 788 | A | 789 | U | 790 | cWW | tSH | cWW | ntsS | tWW | cWW |
| 3 | J3_8CRE_075 | 8CRE | 0.1170 | 0 | CM | SSU rRNA | A | 745 | G | 746 | A | 747 | G | 748 | * | C | 758 | A | 759 | G | 760 | * | U | 769 | C | 770 | G | 771 | A | 772 | A | 773 | U | 774 | cWW | tSH | cWW | tWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AGAG*CAG*UCGAAU | 2 |
| AGAG*CAG*CUGGAU | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GA*A*CGAA | 2 |
| GA*A*UGGA | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 |
|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 |
| Status | Updated, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (3)
- Basepair signature
- cWW-tSH-F-F-cWW-F-tWW-cWW
- Heat map statistics
- Min 0.12 | Avg 0.12 | Max 0.25
Coloring options: