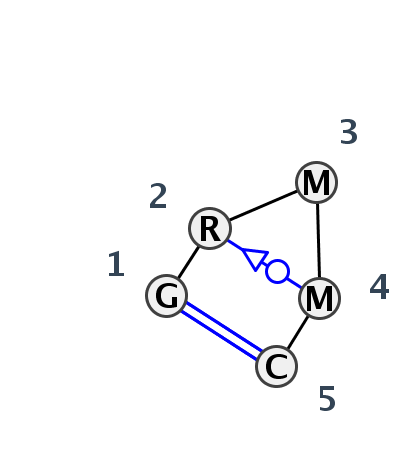
Motif HL_79921.2 Version HL_79921.2 of this group appears in releases 3.80 to 3.82
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 1-5 | 2-3 | 2-4 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_6OL3_001 | 6OL3 | 0.5028 | 2 | Other HL | C | VA RNA | G | 64 | A | 66 | C | 68 | C | 69 | C | 70 | cWW | ntSW | |
| 2 | HL_8T29_001 | 8T29 | 0.1380 | 2 | Fab BL3-6 binding hairpin with tSW | R | RNA (90-MER) | G | 35 | A | 37 | C | 39 | A | 40 | C | 41 | cWW | tSW | |
| 3 | HL_8DP3_001 | 8DP3 | 0.1308 | 2 | Fab BL3-6 binding hairpin with tSW | R | Enterovirus 5' cloverleaf cis-acting replication element | G | 20 | A | 22 | C | 24 | A | 25 | C | 26 | cWW | tSW | |
| 4 | HL_7MLX_002 | 7MLX | 0.1333 | 2 | Fab BL3-6 binding hairpin with tSW | R | RNA (65-MER) | G | 40 | A | 42 | C | 44 | A | 45 | C | 46 | cWW | tSW | |
| 5 | HL_7SZU_001 | 7SZU | 0.1275 | 2 | Fab BL3-6 binding hairpin with tSW | R | RNA aptamer | G | 35 | A | 37 | C | 39 | A | 40 | C | 41 | cWW | tSW | |
| 6 | HL_6X5M_002 | 6X5M | 0.1256 | 2 | Fab BL3-6 binding hairpin with tSW | r | ggPAN RNA (39-MER) | G | 14 | A | 16 | C | 18 | A | 19 | C | 20 | cWW | tSW | |
| 7 | HL_8D29_001 | 8D29 | 0.0981 | 2 | Fab BL3-6 binding hairpin with tSW | C | RNA (34-MER) | G | 13 | A | 15 | C | 17 | A | 18 | C | 19 | cWW | tSW | |
| 8 | HL_8DK7_001 | 8DK7 | 0.0000 | 2 | Fab BL3-6 binding hairpin with tSW | C | RNA (34-MER) | G | 13 | A | 15 | C | 17 | A | 18 | C | 19 | cWW | tSW | |
| 9 | HL_8D29_002 | 8D29 | 0.0634 | 2 | Fab BL3-6 binding hairpin with tSW | F | RNA (34-MER) | G | 13 | A | 15 | C | 17 | A | 18 | C | 19 | cWW | tSW | |
| 10 | HL_8D29_003 | 8D29 | 0.0756 | 2 | Fab BL3-6 binding hairpin with tSW | J | RNA (34-MER) | G | 13 | A | 15 | C | 17 | A | 18 | C | 19 | cWW | tSW | |
| 11 | HL_4KZD_001 | 4KZD | 0.0855 | 2 | Fab BL3-6 binding hairpin with tSW | R | RNA (84-MER) | G | 37 | A | 39 | C | 41 | A | 42 | C | 43 | cWW | tSW | |
| 12 | HL_6DB9_001 | 6DB9 | 0.0748 | 2 | Fab BL3-6 binding hairpin with tSW | R | RNA (60-MER) | G | 23 | A | 25 | C | 27 | A | 28 | C | 29 | cWW | tSW | |
| 13 | HL_3IVK_003 | 3IVK | 0.0944 | 2 | Fab BL3-6 binding hairpin with tSW | M | class I ligase product | G | 59 | A | 61 | C | 63 | A | 64 | C | 65 | cWW | tSW | |
| 14 | HL_7RQB_004 | 7RQB | 0.6311 | 1 | 1A | LSU rRNA | G | 139 | G | 140 | A | 141 | A | 142 | C | 142|||A | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GAAACAC | 12 |
| GAACCCC | 1 |
| GGGAAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AAACA | 12 |
| AACCC | 1 |
| GGAA | 1 |
- Annotations
-
- Fab BL3-6 binding hairpin with tSW (12)
- Other HL (1)
- (1)
- Basepair signature
- cWW-tSW-F
- Heat map statistics
- Min 0.04 | Avg 0.22 | Max 0.93
Coloring options: