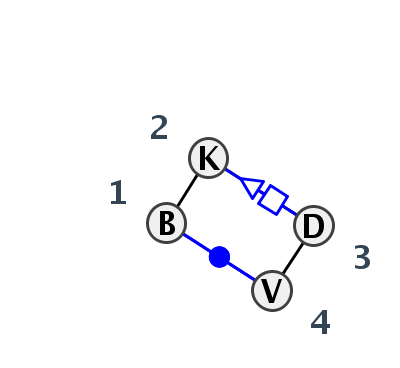
Motif HL_80489.1 Version HL_80489.1 of this group appears in releases 3.77 to 3.79
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 1-4 | 2-3 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_5J7L_180 | 5J7L | 0.8679 | 2 | GNRA variation | DA | LSU rRNA | C | 1868 | G | 1869 | A | 1872 | G | 1873 | cWW | |
| 2 | HL_2CZJ_007 | 2CZJ | 0.6672 | 2 | UNCG variation | H | tmRNA | G | 25 | U | 26 | G | 29 | C | 30 | cWW | |
| 3 | HL_1M5K_003 | 1M5K | 0.6483 | 2 | GNRA | B | Hairpin ribozyme | G | 74 | G | 75 | A | 78 | C | 79 | cWW | ntSH |
| 4 | HL_6CZR_074 | 6CZR | 0.5834 | 2 | GNRA | 1a | SSU rRNA | G | 153 | G | 154 | A | 157 | C | 158 | ncWW | ntSH |
| 5 | HL_7A0S_046 | 7A0S | 0.9089 | 3 | GNRA | X | LSU rRNA | C | 1797 | G | 1798 | A | 1802 | G | 1803 | cWW | tSH |
| 6 | HL_5J7L_026 | 5J7L | 0.7248 | 3 | GNRA | AA | SSU rRNA | U | 1165 | G | 1166 | A | 1170 | A | 1171 | cWW | tSH |
| 7 | HL_8HNT_002 | 8HNT | 0.7442 | 2 | B | sgRNA | U | 103 | G | 104 | A | 107 | G | 108 | cWW | ||
| 8 | HL_3SIU_002 | 3SIU | 0.4816 | 3 | F | U4atac snRNA | C | 40 | G | 41 | U | 44 | G | 46 | cWW | ||
| 9 | HL_6WBR_002 | 6WBR | 0.0000 | 3 | B | RNA (94-MER) | C | 56 | U | 57 | G | 60 | G | 62 |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GGAAAC | 2 |
| CGCAAG | 1 |
| GUUCGC | 1 |
| CGAACAG | 1 |
| UGAUAAA | 1 |
| UGUAAG | 1 |
| CGCAUAG | 1 |
| CUUCGUG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAAA | 2 |
| GCAA | 1 |
| UUCG | 1 |
| GAACA | 1 |
| GAUAA | 1 |
| GUAA | 1 |
| GCAUA | 1 |
| UUCGU | 1 |
- Annotations
-
- GNRA (4)
- (3)
- UNCG variation (1)
- GNRA variation (1)
- Basepair signature
- cWW-tSH
- Heat map statistics
- Min 0.20 | Avg 0.52 | Max 0.91
Coloring options: