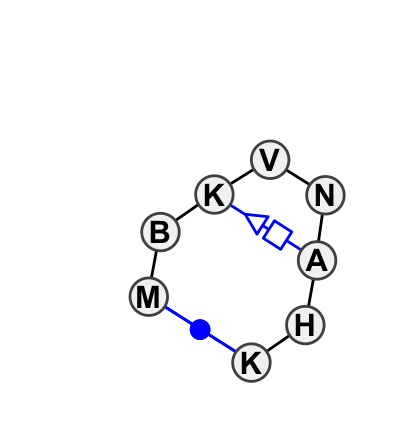
Motif HL_95720.1 Version HL_95720.1 of this group appears in releases 1.5 to 1.18
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 1-8 | 2-7 | 3-6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_1S72_049 | 1S72 | 0.9194 | 0 | GNRA related (H) | 0 | LSU rRNA | C | 1916 | G | 1917 | U | 1918 | A | 1919 | C | 1920 | A | 1921 | A | 1922 | G | 1923 | cWW | ntSH | |
| 2 | HL_1QRS_002 | 1QRS | 0.8997 | 1 | tRNA anticodon loop with synthetase (H) | B | tRNA | A | 31 | U | 32 | U | 33 | C | 34 | U | 35 | A | 37 | U | 38 | U | 39 | cWW | ntSH | |
| 3 | HL_4BPP_027 | 4BPP | 0.2728 | 2 | A | SSU rRNA | A | 1210 | U | 1211 | U | 1212 | G | 1213 | A | 1214 | A | 1216 | C | 1218 | U | 1219 | cWW | ntSH | ||
| 4 | HL_1S72_032 | 1S72 | 0.0000 | 1 | Anticodon loop related (H) | 0 | LSU rRNA | A | 1274 | C | 1275 | U | 1276 | C | 1277 | A | 1278 | A | 1280 | C | 1281 | U | 1282 | cWW | cWB | ntSH |
| 5 | HL_1S72_043 | 1S72 | 0.3060 | 1 | GNRA with tandem sheared (H) | 0 | LSU rRNA | C | 1705 | G | 1706 | G | 1707 | C | 1708 | G | 1709 | A | 1711 | A | 1712 | G | 1713 | cWW | tSH | ntSH |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGUACAAG | 1 |
| AUUCUGAUU | 1 |
| AUUGAGAGCU | 1 |
| ACUCAUACU | 1 |
| CGGCGAAAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GUACAA | 1 |
| UUCUGAU | 1 |
| UUGAGAGC | 1 |
| CUCAUAC | 1 |
| GGCGAAA | 1 |
Release history
| Release | 1.5 | 1.6 | 1.7 | 1.8 | 1.9 | 1.10 | 1.11 | 1.12 | 1.13 | 1.14 | 1.15 | 1.16 | 1.17 | 1.18 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date | 2013-08-03 | 2013-08-31 | 2013-09-28 | 2013-11-04 | 2013-12-07 | 2014-01-04 | 2014-02-01 | 2014-03-01 | 2014-03-29 | 2014-04-27 | 2014-06-15 | 2014-07-26 | 2014-09-06 | 2014-10-09 |
| Status | New id, 2 parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
| Parent motif | Common motif instances | Only in HL_95720.1 | Only in the parent motif |
|---|---|---|---|
| HL_77235.3 Compare | HL_1S72_049, HL_1QRS_002, HL_1S72_032 | HL_1S72_043, HL_4BPP_027 | HL_4A1B_047 |
| HL_52393.1 Compare | HL_1S72_043 | HL_4BPP_027, HL_1S72_049, HL_1QRS_002, HL_1S72_032 |
Children motifs
This motif has no children motifs.- Annotations
-
- tRNA anticodon loop with synthetase (H) (1)
- Anticodon loop related (H) (1)
- GNRA with tandem sheared (H) (1)
- GNRA related (H) (1)
- (1)
- Basepair signature
- cWW-R-tSH-R-R
- Heat map statistics
- Min 0.27 | Avg 0.58 | Max 0.99
Coloring options: