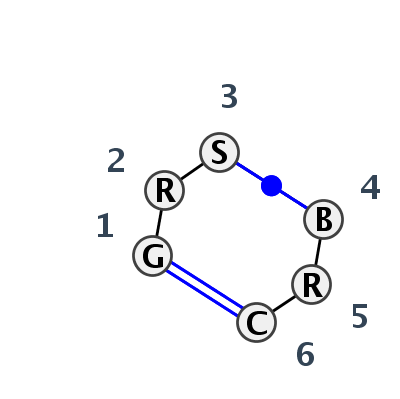
Motif IL_02284.4 Version IL_02284.4 of this group appears in releases 3.77 to 3.79
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | break | 4 | 5 | 6 | 1-6 | 3-4 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_7A0S_100 | 7A0S | 0.2599 | 1 | Decoding loop related | X | LSU rRNA | G | 2818 | G | 2819 | C | 2820 | * | G | 2846 | G | 2847 | C | 2849 | cWW | cWW |
| 2 | IL_4WF9_107 | 4WF9 | 0.0000 | 1 | Decoding loop related (H) | X | LSU rRNA | G | 2863 | A | 2864 | G | 2865 | * | U | 2891 | G | 2892 | C | 2894 | cWW | cWW |
| 3 | IL_5J7L_065 | 5J7L | 0.3477 | 1 | AA | SSU rRNA | G | 1455 | A | 1456 | G | 1457 | * | U | 1445 | A | 1446 | C | 1448 | cWW | cWW | |
| 4 | IL_3NDB_006 | 3NDB | 0.6180 | 1 | M | SRP RNA | G | 152 | A | 153 | G | 154 | * | U | 175 | A | 176 | C | 178 | cWW | cWW | |
| 5 | IL_4M6D_013 | 4M6D | 0.6566 | 0 | H | aptamer | G | 26 | A | 27 | G | 28 | * | C | 39 | A | 40 | C | 41 | cWW | cWW | |
| 6 | IL_4M4O_004 | 4M4O | 0.7091 | 0 | B | RNA (59-MER) | G | 26 | A | 27 | G | 28 | * | C | 39 | A | 40 | C | 41 | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GAG*UAAC | 2 |
| GAG*CAC | 2 |
| GGC*GGAC | 1 |
| GAG*UGAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| A*AA | 2 |
| A*A | 2 |
| G*GA | 1 |
| A*GA | 1 |
- Annotations
-
- (4)
- Decoding loop related (H) (1)
- Decoding loop related (1)
- Basepair signature
- cWW-L-R-cWW
- Heat map statistics
- Min 0.14 | Avg 0.45 | Max 0.80
Coloring options: