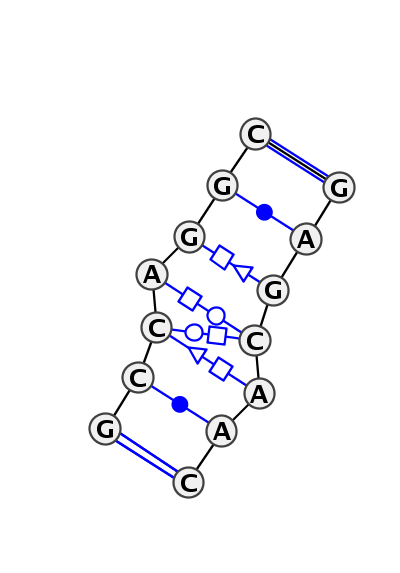
Motif IL_10517.1 Version IL_10517.1 of this group appears in releases 3.46 to 3.61
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 1-14 | 2-13 | 3-11 | 3-12 | 4-11 | 5-10 | 6-9 | 7-8 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_3NDB_008 | 3NDB | 0.0000 | 0 | M | SRP RNA | G | 200 | C | 201 | C | 202 | A | 203 | G | 204 | G | 205 | C | 206 | * | G | 215 | A | 216 | G | 217 | C | 218 | A | 219 | A | 220 | C | 221 | cWW | cWW | ntWH | ntSH | tHW | tHS | cWW | cWW | |
| 2 | IL_3KTW_004 | 3KTW | 0.2181 | 0 | tSH-tHW-tHS with extra cWW pairs | C | SRP RNA | G | 201 | C | 202 | C | 203 | A | 204 | G | 205 | G | 206 | C | 207 | * | G | 216 | A | 217 | G | 218 | C | 219 | A | 220 | A | 221 | C | 222 | cWW | ntWH | tSH | tHW | ntHS | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GCCAGGC*GAGCAAC | 2 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CCAGG*AGCAA | 2 |
Release history
| Release | 3.46 | 3.47 | 3.48 | 3.49 | 3.50 | 3.51 | 3.52 | 3.53 | 3.54 | 3.55 | 3.56 | 3.57 | 3.58 | 3.59 | 3.60 | 3.61 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date | 2021-06-23 | 2021-07-21 | 2021-08-18 | 2021-09-15 | 2021-10-13 | 2021-11-10 | 2021-12-08 | 2022-01-05 | 2022-02-02 | 2022-03-02 | 2022-03-30 | 2022-04-27 | 2022-05-25 | 2022-06-22 | 2022-07-20 | 2022-08-17 |
| Status | New id, 2 parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- tSH-tHW-tHS with extra cWW pairs (1)
- (1)
- Basepair signature
- cWW-cWW-tSH-tWH-tHW-tHS-cWW-cWW
- Heat map statistics
- Min 0.22 | Avg 0.11 | Max 0.22
Coloring options: