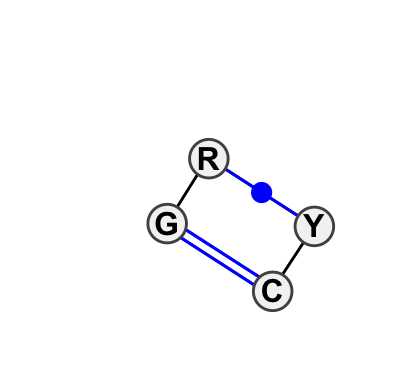
Motif IL_13069.2 Version IL_13069.2 of this group appears in releases 0.7 to 1.0
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | break | 3 | 4 | 1-4 | 2-3 | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_2ZJR_020 | 2ZJR | 0.1468 | 4 | Multiple bulged bases (H) | X | LSU rRNA | G | 788 | A | 790 | * | U | 800 | C | 804 | cWW | cWW |
| 2 | IL_1S72_031 | 1S72 | 0.1054 | 4 | Multiple bulged bases (H) | 0 | LSU rRNA | G | 868 | G | 870 | * | C | 880 | C | 884 | cWW | cWW |
| 3 | IL_3V2F_023 | 3V2F | 0.0944 | 4 | A | LSU rRNA | G | 775 | A | 777 | * | U | 787 | C | 791 | cWW | cWW | |
| 4 | IL_2QBG_026 | 2QBG | 0.0000 | 4 | B | LSU rRNA | G | 775 | G | 777 | * | C | 787 | C | 791 | cWW | cWW | |
| 5 | IL_3U5H_035 | 3U5H | 0.1712 | 4 | 5 | LSU rRNA | G | 907 | G | 909 | * | U | 919 | C | 923 | cWW | cWW | |
| 6 | IL_4A1B_040 | 4A1B | 0.1860 | 4 | 1 | LSU rRNA | G | 932 | G | 934 | * | U | 944 | C | 948 | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GGA*UAACC | 2 |
| GGG*UAAUC | 2 |
| GGG*CCAUC | 1 |
| GGG*CAAUC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| G*AAU | 3 |
| G*AAC | 2 |
| G*CAU | 1 |
Release history
| Release | 0.7 | 0.8 | 0.9 | 0.10 | 0.11 | 0.12 | 1.0 |
|---|---|---|---|---|---|---|---|
| Date | 2013-02-17 | 2013-02-17 | 2013-02-17 | 2013-02-17 | 2013-02-17 | 2013-02-17 | 2013-03-04 |
| Status | Updated, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
| Parent motif | Common motif instances | Only in IL_13069.2 | Only in the parent motif |
|---|---|---|---|
| IL_13069.1 Compare | IL_4A1B_040, IL_3U5H_035, IL_2QBG_026, IL_1S72_031, IL_2ZJR_020 | IL_3V2F_023 | IL_3UXR_023 |
Children motifs
| Child motif | Common motif instances | Only in IL_13069.2 | Only in the child motif |
|---|---|---|---|
| IL_13069.3 Compare | IL_4A1B_040, IL_3U5H_035, IL_2QBG_026, IL_1S72_031, IL_3V2F_023 | IL_2ZJR_020 | IL_4IOA_020 |
- Annotations
-
- (4)
- Multiple bulged bases (H) (2)
- Basepair signature
- cWW-cWW
- Heat map statistics
- Min 0.07 | Avg 0.14 | Max 0.27
Coloring options: