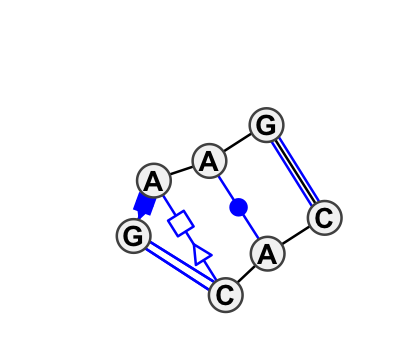
Motif IL_20017.1 Version IL_20017.1 of this group appears in releases 0.4 to 0.4
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | break | 5 | 6 | 7 | 1-7 | 2-7 | 3-6 | 4-5 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_3MUR_001 | 3MUR | 0.7502 | 1 | R | Cyclic di-GMP-I riboswitch | G | 94 | A | 95 | G | 97 | G | 98 | * | C | 11 | A | 12 | C | 13 | cWW | cWW | cWW | |
| 2 | IL_3BNL_003 | 3BNL | 0.0000 | 0 | A+B | A site of bacterial ribosome | G | 15 | A | 16 | A | 17 | G | 18 | * | C | 6 | A | 7 | C | 8 | cWW | cWw | cWW | |
| 3 | IL_3F1H_030 | 3F1H | 0.3894 | 0 | A | LSU rRNA | A | 899 | A | 900 | A | 901 | C | 902 | * | G | 875 | C | 876 | U | 877 | cWW | ntHS | ncWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GAUGG*CAC | 1 |
| GAAG*CAC | 1 |
| AAAC*GCU | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AUG*A | 1 |
| AA*A | 1 |
| AA*C | 1 |
Release history
| Release | 0.4 |
|---|---|
| Date | 2011-12-08 |
| Status | > 2 parents |
Parent motifs
| Parent motif | Common motif instances | Only in IL_20017.1 | Only in the parent motif |
|---|---|---|---|
| IL_00449.1 Compare | IL_3F1H_030 | IL_3MUR_001, IL_3BNL_003 | |
| IL_34635.2 Compare | IL_3BNL_003 | IL_3MUR_001, IL_3F1H_030 | IL_3F1H_106, IL_1LNG_001 |
| IL_72861.1 Compare | IL_3MUR_001 | IL_3F1H_030, IL_3BNL_003 |
Children motifs
| Child motif | Common motif instances | Only in IL_20017.1 | Only in the child motif |
|---|---|---|---|
| IL_09107.1 Compare | IL_3MUR_001 | IL_3F1H_030, IL_3BNL_003 | |
| IL_69110.1 Compare | IL_3BNL_003 | IL_3MUR_001, IL_3F1H_030 | IL_3PYO_030 |
Coloring options: