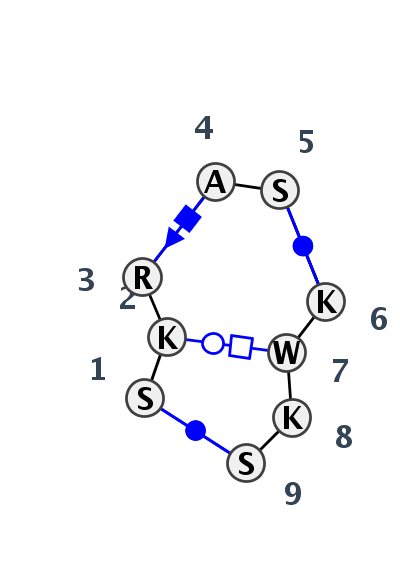
Motif IL_44312.1 Version IL_44312.1 of this group appears in releases 3.77 to 3.83
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 1-9 | 2-7 | 2-8 | 3-4 | 3-6 | 4-7 | 4-8 | 5-6 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_3IGI_012 | 3IGI | 0.2299 | 0 | Receptor of 11-nt loop-receptor motif | A | Group II catalytic intron D1-D4-3 | C | 346 | U | 347 | A | 348 | A | 349 | G | 350 | * | U | 332 | A | 333 | U | 334 | G | 335 | cWW | tWH | cSH | ncWW | cWW | |||
| 2 | IL_1U6B_003 | 1U6B | 0.0000 | 0 | Receptor of 11-nt loop-receptor motif | B | 197-MER | C | 61 | U | 62 | A | 63 | A | 64 | G | 65 | * | U | 80 | A | 81 | U | 82 | G | 83 | cWW | tWH | cSH | ncWW | cWW | |||
| 3 | IL_3KTW_003 | 3KTW | 0.8054 | 2 | C | SRP RNA | G | 193 | G | 194 | G | 195 | A | 196 | C | 199 | * | G | 224 | U | 225 | G | 226 | C | 227 | cWW | tSH | ntWH | ntHW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CUAAG*UAUG | 2 |
| GGGAACC*GUGC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UAA*AU | 2 |
| GGAAC*UG | 1 |
Release history
| Release | 3.77 | 3.78 | 3.79 | 3.80 | 3.81 | 3.82 | 3.83 |
|---|---|---|---|---|---|---|---|
| Date | 2023-12-06 | 2024-01-03 | 2024-01-03 | 2024-01-31 | 2024-02-28 | 2024-03-27 | 2024-04-24 |
| Status | New id, 2 parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- Receptor of 11-nt loop-receptor motif (2)
- (1)
- Basepair signature
- cWW-tWH-R-cSH-cWW
- Heat map statistics
- Min 0.23 | Avg 0.42 | Max 0.85
Coloring options: