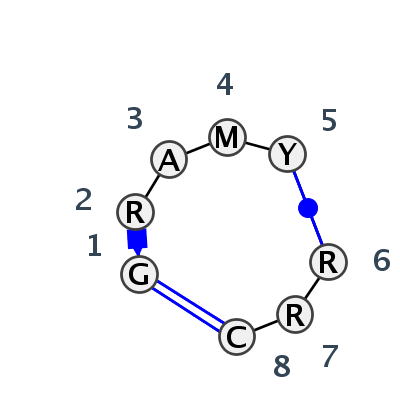
Motif IL_47973.1 Version IL_47973.1 of this group appears in releases 3.77 to 3.79
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 1-2 | 1-8 | 2-8 | 3-8 | 5-6 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_5J7L_013 | 5J7L | 0.5243 | 0 | AA | SSU rRNA | G | 203 | G | 204 | A | 205 | C | 206 | C | 207 | * | G | 212 | G | 213 | C | 214 | cWW | cWW | |||
| 2 | IL_4YAZ_005 | 4YAZ | 0.0000 | 1 | R | Cyclic di-GMP-I riboswitch | G | 77 | A | 78 | A | 79 | A | 80 | U | 81 | * | A | 4 | A | 6 | C | 7 | cSH | cWW | ntHS | cWS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GGACC*GGC | 1 |
| GAAAU*ACAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAC*G | 1 |
| AAA*CA | 1 |
- Annotations
-
- (2)
- Basepair signature
- cWW-cSH-R-L-L-cWW
- Heat map statistics
- Min 0.52 | Avg 0.26 | Max 0.52
Coloring options: