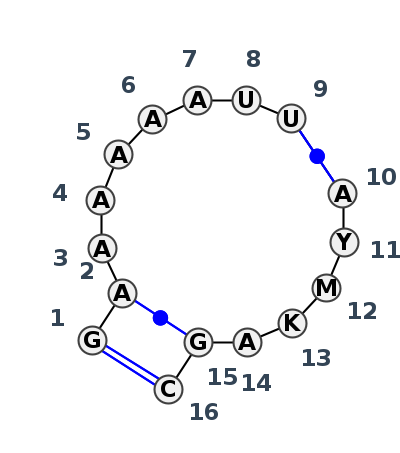
Motif IL_59049.2 Version IL_59049.2 of this group appears in releases 4.0 to 4.6
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | break | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 1-16 | 2-15 | 3-13 | 9-10 | 13-14 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_8P9A_414 | 8P9A | 0.4879 | 1 | sR | SSU rRNA | G | 751 | A | 752 | A | 753 | A | 754 | A | 755 | A | 756 | A | 757 | U | 758 | U | 759 | * | A | 791 | U | 792 | A | 793 | U | 794 | A | 796 | G | 797 | C | 798 | cWW | cWW | cWW | ||
| 2 | IL_8CRE_430 | 8CRE | 0.0000 | 1 | CM | SSU rRNA | G | 736 | A | 737 | A | 738 | A | 739 | A | 740 | A | 741 | A | 742 | U | 743 | U | 744 | * | A | 775 | U | 776 | A | 777 | U | 778 | A | 780 | G | 781 | C | 782 | cWW | cWW | ncWW | cWW | ncSH |
| 3 | IL_8GLP_330 | 8GLP | 0.4028 | 1 | S2 | SSU rRNA | G | 807 | A | 808 | A | 809 | A | 810 | A | 811 | A | 812 | A | 813 | PSU | 814 | PSU | 815 | * | A | 849 | C | 850 | C | 851 | G | 852 | A | 854 | G | 855 | C | 856 | cWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GAAAAAAUU*AUAUUAGC | 2 |
| GAAAAAA(PSU)(PSU)*ACCGCAGC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AAAAAAU*UAUUAG | 2 |
| AAAAAA(PSU)*CCGCAG | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 |
|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 |
| Status | Updated, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (3)
- Basepair signature
- cWW-cWW-L-R-L-R-L-R-L-R-L-cWW-L
- Heat map statistics
- Min 0.40 | Avg 0.33 | Max 0.59
Coloring options: