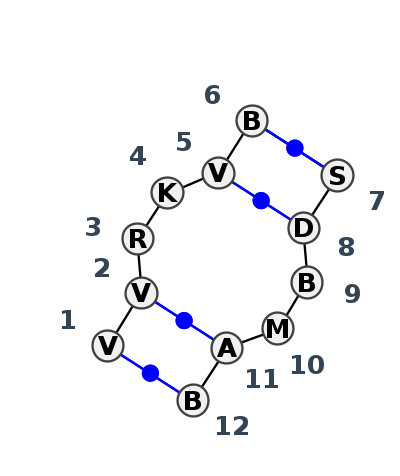
Motif IL_71294.1 Version IL_71294.1 of this group appears in releases 3.89 to 3.89
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | 10 | 11 | 12 | 1-12 | 2-11 | 3-10 | 4-9 | 5-8 | 6-7 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_4N0T_001 | 4N0T | 0.5430 | 0 | Double sheared; A in syn; two near cWW pairs | B | U6 snRNA | A | 95 | G | 96 | A | 97 | G | 98 | A | 99 | C | 100 | * | G | 31 | U | 32 | C | 33 | A | 34 | A | 35 | U | 36 | cWW | ncWW | cWW | |||
| 2 | IL_1L9A_008 | 1L9A | 0.0000 | 0 | tSH-tHW-tHS-cWW | B | SRP RNA | C | 190 | C | 191 | A | 192 | G | 193 | G | 194 | U | 195 | * | G | 204 | A | 205 | G | 206 | C | 207 | A | 208 | G | 209 | cWW | ncWW | tHW | cBW | cWW | cWW |
| 3 | IL_6CZR_369 | 6CZR | 0.4289 | 1 | 1a | SSU rRNA | G | 1469 | A | 1470 | G | 1472 | U | 1473 | C | 1474 | G | 1475 | * | 5MC | 1387 | G | 1388 | U | 1389 | 5MC | 1390 | A | 1391 | C | 1392 | cWW | cwW | ncWW | ncWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AGAGAC*GUCAAU | 1 |
| CCAGGU*GAGCAG | 1 |
| GAAGUCG*(5MC)GU(5MC)AC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAGA*UCAA | 1 |
| CAGG*AGCA | 1 |
| AAGUC*GU(5MC)A | 1 |
Release history
| Release | 3.89 |
|---|---|
| Date | 2024-10-09 |
| Status | New id, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- tSH-tHW-tHS-cWW (1)
- Double sheared; A in syn; two near cWW pairs (1)
- (1)
- Basepair signature
- cWW-cWW-L-R-L-R-cWW-cWW
- Heat map statistics
- Min 0.43 | Avg 0.35 | Max 0.60
Coloring options: