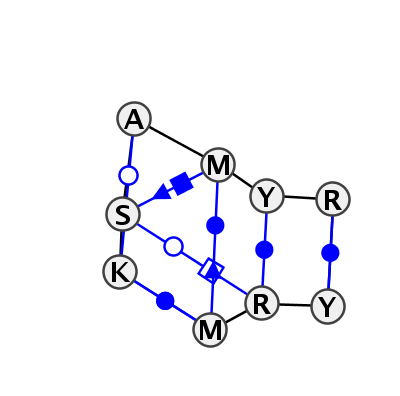
Motif IL_71655.1 Version IL_71655.1 of this group appears in releases 2.0 to 2.0
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | 1-3 | 1-9 | 2-4 | 2-5 | 2-8 | 4-9 | 5-8 | 6-7 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_3J9W_094 | 3J9W | 0.2612 | 2 | BA | LSU rRNA | G | 911 | C | 912 | A | 913 | C | 914 | U | 915 | G | 916 | * | C | 955 | A | 956 | C | 959 | tSW | cWW | ncSH | tWH | cWW | cWW | ||
| 2 | IL_4V9F_035 | 4V9F | 0.0000 | 3 | 0 | LSU rRNA | G | 958 | C | 959 | A | 961 | C | 962 | C | 963 | G | 964 | * | C | 1004 | A | 1005 | C | 1008 | tSW | cWW | cSH | ntSW | tWH | cWS | cWW | cWW |
| 3 | IL_5FDU_052 | 5FDU | 0.3502 | 1 | 1A | LSU rRNA | G | 1365 | C | 1366 | A | 1367 | A | 1368 | U | 1369 | G | 1370 | * | C | 1376 | A | 1377 | C | 1379 | cWW | tWH | cWS | cWW | cWW | |||
| 4 | IL_5AJ0_099 | 5AJ0 | 0.5465 | 1 | A2 | LSU rRNA | U | 2394 | G | 2395 | A | 2396 | A | 2397 | C | 2398 | A | 2399 | * | U | 2405 | G | 2406 | A | 2408 | cWW | cWS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GCACUG*CAAAC | 1 |
| GCGACCG*CAAAC | 1 |
| GCAAUG*CAGC | 1 |
| UGAACA*UGAA | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CACU*AAA | 1 |
| CGACC*AAA | 1 |
| CAAU*AG | 1 |
| GAAC*GA | 1 |
Release history
| Release | 2.0 |
|---|---|
| Date | 2017-04-24 |
| Status | New id, no parents |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (4)
- Basepair signature
- cWW-tSW-tWH-cSH-cWS-cWW-cWW
- Heat map statistics
- Min 0.26 | Avg 0.33 | Max 0.62
Coloring options: