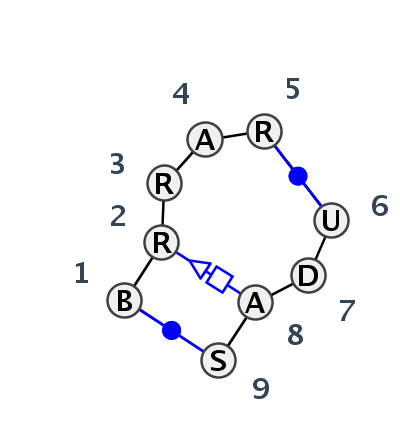
Motif IL_77691.1 Version IL_77691.1 of this group appears in releases 3.80 to 3.83
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 1-9 | 2-8 | 3-7 | 5-6 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_4V88_390 | 4V88 | 0.4445 | 2 | A6 | SSU rRNA | U | 82 | G | 83 | A | 84 | A | 85 | A | 86 | * | U | 64 | A | 65 | A | 67 | G | 69 | cWW | tSH | cWW | ||
| 2 | IL_8C3A_398 | 8C3A | 0.4527 | 2 | CM | SSU rRNA | U | 82 | G | 83 | A | 84 | A | 85 | A | 86 | * | U | 64 | A | 65 | A | 67 | G | 69 | cWW | tSH | cWW | ||
| 3 | IL_2R8S_002 | 2R8S | 0.2798 | 0 | UAA/GAN related | R | P4-P6 RNA RIBOZYME DOMAIN | G | 112 | A | 113 | A | 114 | A | 115 | G | 116 | * | U | 205 | A | 206 | A | 207 | C | 208 | cWW | tSH | tHS | cWW |
| 4 | IL_3FS0_001 | 3FS0 | 0.2562 | 0 | UAA/GAN related | B+A | RNA (11-mer) | C | -1 | G | 1 | G | 2 | A | 3 | A | 4 | * | U | 46 | G | 47 | A | 48 | G | 49 | cWW | tSH | ntHh | cWW |
| 5 | IL_5J7L_317 | 5J7L | 0.1438 | 0 | UAA/GAN related | DA | LSU rRNA | C | 1874 | G | 1875 | A | 1876 | A | 1877 | G | 1878 | * | U | 1864 | U | 1865 | A | 1866 | G | 1867 | cWW | tSH | cWW | |
| 6 | IL_4WF9_075 | 4WF9 | 0.0000 | 0 | UAA/GAN variation | X | LSU rRNA | C | 1901 | G | 1902 | A | 1903 | A | 1904 | G | 1905 | * | U | 1891 | U | 1892 | A | 1893 | G | 1894 | cWW | ntSH | ntHW | cWW |
| 7 | IL_3FTM_002 | 3FTM | 0.3370 | 0 | UAA/GAN related | D+C | RNA (12-mer) | C | -1 | G | 1 | G | 2 | A | 3 | A | 4 | * | U | 46 | G | 47 | A | 48 | G | 49 | cWW | ntSH | cWW | |
| 8 | IL_3IGI_009 | 3IGI | 0.4531 | 0 | UAA/GAN related | A | Group II catalytic intron D1-D4-3 | C | 190 | A | 191 | A | 192 | A | 193 | G | 194 | * | U | 172 | G | 173 | A | 174 | G | 175 | cWW | tSH | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UGAAA*UAUAAG | 2 |
| CGGAA*UGAG | 2 |
| CGAAG*UUAG | 2 |
| GAAAG*UAAC | 1 |
| CAAAG*UGAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAA*AUAA | 2 |
| GGA*GA | 2 |
| GAA*UA | 2 |
| AAA*AA | 1 |
| AAA*GA | 1 |
- Annotations
-
- UAA/GAN related (5)
- (2)
- UAA/GAN variation (1)
- Basepair signature
- cWW-tSH-L-R-L-cWW
- Heat map statistics
- Min 0.14 | Avg 0.31 | Max 0.51
Coloring options: