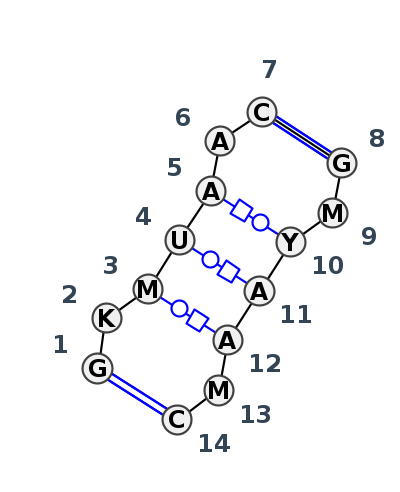
Motif IL_80604.4 Version IL_80604.4 of this group appears in releases 3.91 to 3.94
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 1-14 | 2-13 | 3-12 | 3-13 | 4-11 | 5-10 | 6-9 | 7-8 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_6CZR_121 | 6CZR | 0.4640 | 1 | tWH-tWH-tHW-tHW | 1a | SSU rRNA | G | 162 | G | 163 | C | 164 | U | 165 | A | 166 | A | 167 | C | 169 | * | G | 143 | A | 144 | C | 145 | A | 146 | A | 147 | C | 148 | C | 149 | ncWW | ncBW | ntWH | ntHW | ncBW | cWW | ||
| 2 | IL_5J7L_012 | 5J7L | 0.1705 | 1 | tWH-tWH-tHW-tHW | AA | SSU rRNA | G | 147 | G | 148 | A | 149 | U | 150 | A | 151 | A | 152 | C | 153 | * | G | 168 | C | 169 | U | 170 | A | 171 | A | 172 | A | 174 | C | 175 | cWW | cWW | tWH | tWH | tHW | cWW | ||
| 3 | IL_8P9A_383 | 8P9A | 0.1088 | 1 | tWH-tWH-tHW-tHW (H) | sR | SSU rRNA | G | 143 | U | 144 | A | 145 | U | 146 | A | 147 | A | 148 | C | 149 | * | G | 165 | C | 166 | U | 167 | A | 168 | A | 169 | A | 171 | C | 172 | cWW | tWH | tWH | tHW | tHW | cWW | ||
| 4 | IL_8C3A_402 | 8C3A | 0.0000 | 1 | tWH-tWH-tHW-tHW (H) | CM | SSU rRNA | G | 141 | G | 142 | A | 143 | U | 144 | A | 145 | A | 146 | C | 147 | * | G | 163 | C | 164 | U | 165 | A | 166 | A | 167 | A | 169 | C | 170 | cWW | cWW | tWH | tHW | tHW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GGAUAAC*GCUAAUAC | 2 |
| GGCUAAUC*GACAACC | 1 |
| GUAUAAC*GCUAAUAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAUAA*CUAAUA | 2 |
| GCUAAU*ACAAC | 1 |
| UAUAA*CUAAUA | 1 |
- Annotations
-
- tWH-tWH-tHW-tHW (2)
- tWH-tWH-tHW-tHW (H) (2)
- Basepair signature
- cWW-L-R-tWH-tWH-tHW-L-R-cWW
- Heat map statistics
- Min 0.11 | Avg 0.23 | Max 0.48
Coloring options: