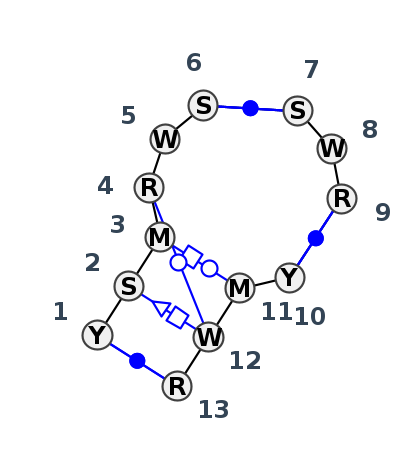
Motif J3_10154.2 Version J3_10154.2 of this group appears in releases 3.89 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | break | 10 | 11 | 12 | 13 | 1-13 | 2-12 | 3-11 | 4-8 | 4-12 | 6-7 | 9-10 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_5J7L_042 | 5J7L | 0.4987 | 1 | DA | LSU rRNA | C | 1605 | C | 1606 | C | 1607 | A | 1608 | A | 1610 | C | 1611 | * | G | 1620 | U | 1621 | G | 1622 | * | C | 1306 | A | 1307 | A | 1308 | G | 1309 | cWW | ntSH | tHW | tWW | cWW | cWW | |
| 2 | J3_8C3A_006 | 8C3A | 0.0000 | 1 | 1 | LSU rRNA | U | 1475 | G | 1476 | A | 1477 | G | 1479 | U | 1480 | G | 1481 | * | C | 1853 | A | 1854 | A | 1855 | * | U | 1867 | C | 1868 | U | 1869 | A | 1870 | cWW | tSH | tHW | tWW | cWW | cWW | |
| 3 | J3_5TBW_008 | 5TBW | 0.0823 | 1 | 1 | LSU rRNA | U | 1479 | G | 1480 | A | 1481 | G | 1483 | U | 1484 | G | 1485 | * | C | 1857 | A | 1858 | A | 1859 | * | U | 1871 | C | 1872 | U | 1873 | A | 1874 | cWW | tSH | tHW | tWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UGAAGUG*CAA*UCUA | 2 |
| CAAG*CCCAAAC*GUG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAAGU*A*CU | 2 |
| AA*CCAAA*U | 1 |
- Annotations
-
- (3)
- Basepair signature
- cWW-tWW-tHW-cWW-tSH-cWW-cWW-F-F
- Heat map statistics
- Min 0.08 | Avg 0.24 | Max 0.51
Coloring options: