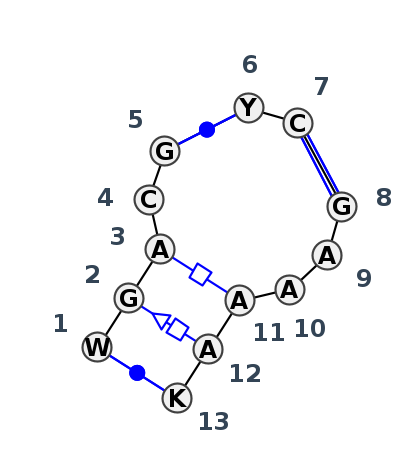
Motif J3_12393.2 Version J3_12393.2 of this group appears in releases 3.95 to 3.99
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 13 | 1-13 | 2-12 | 3-11 | 5-6 | 7-8 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_8P9A_058 | 8P9A | 0.0922 | 0 | AR | LSU rRNA | A | 1654 | G | 1655 | A | 1656 | C | 1657 | G | 1658 | * | C | 1791 | C | 1792 | * | G | 1796 | A | 1797 | A | 1798 | A | 1799 | A | 1800 | U | 1801 | cWW | tSH | tHH | cWW | cWW |
| 2 | J3_8CRE_018 | 8CRE | 0.0000 | 0 | 1 | LSU rRNA | A | 1650 | G | 1651 | A | 1652 | C | 1653 | G | 1654 | * | U | 1787 | C | 1788 | * | G | 1792 | A | 1793 | A | 1794 | A | 1795 | A | 1796 | U | 1797 | cWW | tSH | tHH | cWW | cWW |
| 3 | J3_4V9F_017 | 4V9F | 0.1147 | 0 | 0 | LSU rRNA | U | 1531 | G | 1532 | A | 1533 | C | 1534 | G | 1535 | * | C | 1650 | C | 1651 | * | G | 1655 | A | 1656 | A | 1657 | A | 1658 | A | 1659 | G | 1660 | cWW | tSH | tHH | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AGACG*CC*GAAAAU | 1 |
| AGACG*UC*GAAAAU | 1 |
| UGACG*CC*GAAAAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAC**AAAA | 3 |
- Annotations
-
- (3)
- Basepair signature
- cWW-tSH-tHH-F-F-cWW-F-cWW
- Heat map statistics
- Min 0.09 | Avg 0.08 | Max 0.14
Coloring options: