Motif J3_42221.1 Version J3_42221.1 of this group appears in releases 3.87 to 3.87
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | 10 | 11 | break | 12 | 13 | 1-13 | 2-10 | 3-9 | 5-8 | 6-7 | 9-11 | 9-12 | 11-12 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
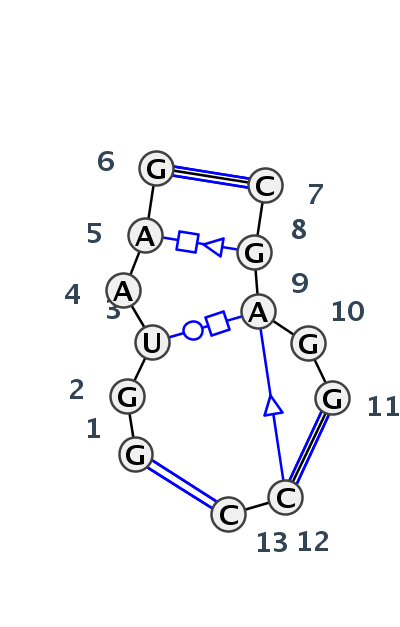
| 1 | J3_8VTW_009 | 8VTW | 0.2880 | 1 | 1A | LSU rRNA | G | 2805 | G | 2807 | U | 2808 | A | 2809 | A | 2810 | G | 2811 | * | C | 2889 | G | 2891 | A | 2892 | G | 2893 | G | 2894 | * | C | 2789 | C | 2791 | cWW | ncWB | tWH | tHS | cWW | ntSs | cWW | |
| 2 | J3_7A0S_012 | 7A0S | 0.0000 | 1 | X | LSU rRNA | G | 2781 | G | 2782 | U | 2783 | A | 2784 | A | 2785 | G | 2786 | * | C | 2864 | G | 2865 | A | 2866 | G | 2867 | G | 2868 | * | C | 2769 | C | 2771 | cWW | ntWW | tWH | tHS | cWW | ncSs | tSs | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CAC*GGUAAG*CGAGG | 2 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| A*GUAA*GAG | 2 |
Release history
| Release | 3.87 |
|---|---|
| Date | 2024-08-14 |
| Status | New id, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (2)
- Basepair signature
- cWW-F-cWW-tSS-tWH-F-F-tHS-cWW
- Heat map statistics
- Min 0.29 | Avg 0.14 | Max 0.29
Coloring options: