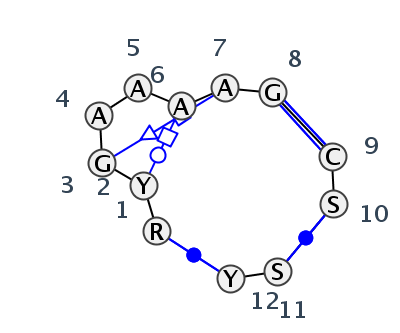
Motif J3_65070.2 Version J3_65070.2 of this group appears in releases 3.48 to 3.53
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | break | 9 | 10 | break | 11 | 12 | 1-12 | 2-6 | 3-7 | 8-9 | 10-11 | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_5TBW_002 | 5TBW | 0.1636 | 0 | 1+4 | LSU rRNA + 5.8S rRNA | A | 369 | U | 370 | G | 371 | A | 372 | A | 373 | A | 374 | A | 375 | G | 376 | * | C | 403 | G | 404 | * | C | 19 | U | 20 | cWW | tWH | tSH | cWW | cWW |
| 2 | J3_5J7L_036 | 5J7L | 0.1156 | 0 | DA | LSU rRNA | G | 474 | C | 475 | G | 476 | A | 477 | A | 478 | A | 479 | A | 480 | G | 481 | * | C | 509 | C | 510 | * | G | 30 | C | 31 | cWW | ntWH | tSH | cWW | cWW |
| 3 | J3_7A0S_001 | 7A0S | 0.0000 | 0 | X | LSU rRNA | G | 485 | U | 486 | G | 487 | A | 488 | A | 489 | A | 490 | A | 491 | G | 492 | * | C | 519 | C | 520 | * | G | 30 | C | 31 | cWW | tWH | ntSH | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| AUGAAAAG*CG*CU | 1 |
| GC*GCGAAAAG*CC | 1 |
| GC*GUGAAAAG*CC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UGAAAA** | 1 |
| *CGAAAA* | 1 |
| *UGAAAA* | 1 |
- Annotations
-
- (3)
- Basepair signature
- cWW-tWH-cWW-tSH-F-cWW-F
- Heat map statistics
- Min 0.12 | Avg 0.09 | Max 0.16
Coloring options: