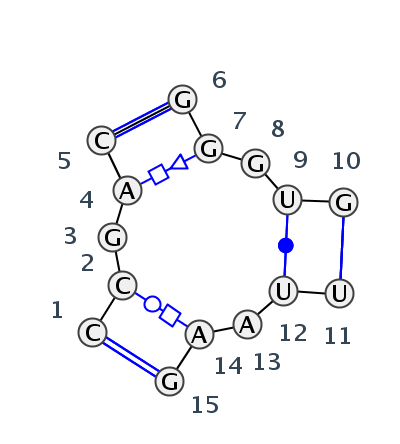
Motif J3_75993.1 Version J3_75993.1 of this group appears in releases 3.80 to 3.87
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 10 | break | 11 | 12 | 13 | 14 | 15 | 1-15 | 2-14 | 4-7 | 5-6 | 8-13 | 9-12 | 10-11 | 11-14 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_8C3A_041 | 8C3A | 0.0000 | 3 | CM | SSU rRNA | C | 1200 | C | 1201 | G | 1203 | A | 1204 | C | 1205 | * | G | 1248 | G | 1249 | G | 1250 | U | 1251 | G | 1252 | * | U | 1428 | U | 1429 | A | 1430 | A | 1432 | G | 1434 | cWW | tWH | tHS | cWW | ncHH | cWW | cWW | ncSS | |
| 2 | J3_4V88_038 | 4V88 | 0.1249 | 3 | A6 | SSU rRNA | C | 1215 | C | 1216 | G | 1218 | A | 1219 | C | 1220 | * | G | 1263 | G | 1264 | G | 1265 | U | 1266 | G | 1267 | * | U | 1442 | U | 1443 | A | 1444 | A | 1446 | G | 1448 | cWW | tWH | tHS | cWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CCAGAC*GGGUG*UUAGACG | 2 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CAGA*GGU*UAGAC | 2 |
Release history
| Release | 3.80 | 3.81 | 3.82 | 3.83 | 3.84 | 3.85 | 3.86 | 3.87 |
|---|---|---|---|---|---|---|---|---|
| Date | 2024-01-31 | 2024-02-28 | 2024-03-27 | 2024-04-24 | 2024-05-22 | 2024-06-19 | 2024-07-17 | 2024-08-14 |
| Status | New id, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
- Basepair signature
- cWW-tWH-F-F-tHS-cWW-cWW-cWW-F
- Heat map statistics
- Min 0.12 | Avg 0.06 | Max 0.12
Coloring options: