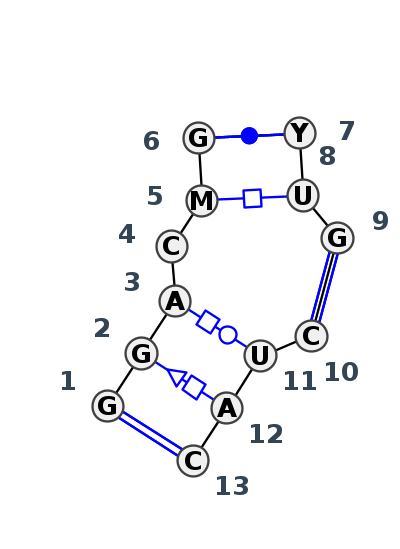
Motif J3_89368.4 Version J3_89368.4 of this group appears in releases 3.87 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | break | 10 | 11 | 12 | 13 | 1-13 | 2-12 | 3-11 | 4-13 | 5-8 | 6-7 | 9-10 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J3_7A0S_009 | 7A0S | 0.1508 | 1 | X | LSU rRNA | G | 2202 | G | 2203 | A | 2204 | C | 2205 | C | 2206 | G | 2207 | * | U | 2074 | U | 2075 | G | 2076 | * | C | 2179 | U | 2180 | A | 2182 | C | 2183 | cWW | tSH | tHW | tHH | cWW | cWW | ||
| 2 | J3_5J7L_044 | 5J7L | 0.0000 | 1 | Kink-turn 3-way junction | DA | LSU rRNA | G | 2223 | G | 2224 | A | 2225 | C | 2226 | A | 2227 | G | 2228 | * | C | 2091 | U | 2092 | G | 2093 | * | C | 2196 | U | 2197 | A | 2199 | C | 2200 | cWW | tSH | tHW | ntWS | tHH | cWW | cWW |
| 3 | J3_8VTW_006 | 8VTW | 0.1238 | 1 | 1A | LSU rRNA | G | 2223 | G | 2224 | A | 2225 | C | 2226 | A | 2227 | G | 2228 | * | U | 2091 | U | 2092 | G | 2093 | * | C | 2196 | U | 2197 | A | 2199 | C | 2200 | cWW | tSH | tHW | tHH | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UUG*CUAAC*GGACCG | 1 |
| CUG*CUAAC*GGACAG | 1 |
| UUG*CUAAC*GGACAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| U*UAA*GACA | 2 |
| U*UAA*GACC | 1 |
- Annotations
-
- (2)
- Kink-turn 3-way junction (1)
- Basepair signature
- cWW-tSH-tHW-F-cWW-tHH-cWW
- Heat map statistics
- Min 0.12 | Avg 0.09 | Max 0.15
Coloring options: