Motif J3_97971.1 Version J3_97971.1 of this group appears in releases 3.88 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | break | 10 | 11 | 12 | 13 | 14 | 1-14 | 3-12 | 4-5 | 4-11 | 5-7 | 5-8 | 7-8 | 9-10 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
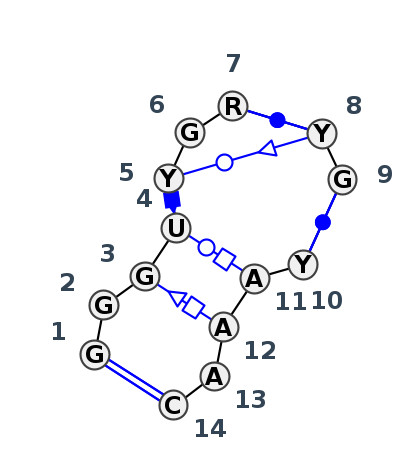
| 1 | J3_7A0S_003 | 7A0S | 0.1995 | 1 | X | LSU rRNA | G | 269 | G | 270 | G | 271 | U | 272 | U | 273 | G | 274 | A | 276 | * | U | 382 | G | 383 | * | C | 246 | A | 247 | A | 248 | A | 249 | C | 250 | cWW | tSH | ncSH | tWH | ntWS | cWW | cWW | |
| 2 | J3_4WF9_022 | 4WF9 | 0.0000 | 1 | X | LSU rRNA | G | 295 | G | 296 | G | 297 | U | 298 | U | 299 | G | 300 | A | 302 | * | U | 415 | G | 416 | * | C | 272 | A | 273 | A | 274 | A | 275 | C | 276 | cWW | ntSH | cSH | tWH | tWS | ncWW | cWW | |
| 3 | J3_8VTW_040 | 8VTW | 0.1564 | 1 | 1A | LSU rRNA | G | 271|||V | G | 271|||W | G | 271|||X | U | 271|||Y | C | 271|||Z | G | 272 | G | 272|||B | * | C | 366 | G | 370 | * | U | 269 | A | 270 | A | 271 | A | 271|||A | C | 271|||B | cWW | tSH | tWH | ncSs | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CAAAC*GGGUUGUA*UG | 2 |
| UAAAC*GGGUCGUG*CG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AAA*GGUUGU* | 2 |
| AAA*GGUCGU* | 1 |
- Annotations
-
- (3)
- Basepair signature
- cWW-F-F-tSH-cSH-tWH-tWS-cWW-F-cWW
- Heat map statistics
- Min 0.16 | Avg 0.13 | Max 0.22
Coloring options: