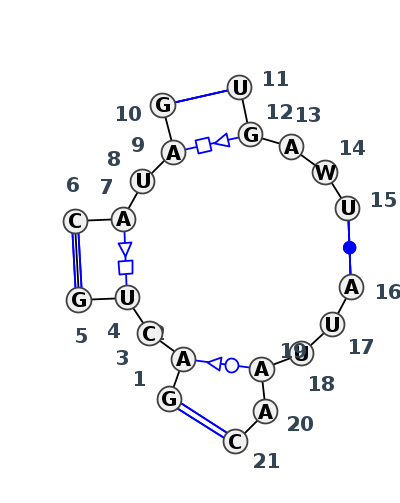
Motif J4_07509.1 Version J4_07509.1 of this group appears in releases 3.91 to 3.94
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 10 | break | 11 | 12 | 13 | 14 | 15 | break | 16 | 17 | 18 | 19 | 20 | 21 | 1-21 | 2-19 | 3-8 | 4-7 | 5-6 | 9-12 | 10-11 | 13-18 | 15-16 | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J4_8P9A_017 | 8P9A | 0.0000 | 0 | AR | LSU rRNA | G | 2130 | A | 2131 | C | 2132 | U | 2133 | G | 2134 | * | C | 2146 | A | 2147 | U | 2148 | A | 2149 | G | 2150 | * | U | 2186 | G | 2187 | A | 2188 | U | 2189 | U | 2190 | * | A | 2317 | U | 2318 | U | 2319 | A | 2320 | A | 2321 | C | 2322 | cWW | tSW | ntHSa | tHS | cWW | tHS | cWW | ntHS | cWW |
| 2 | J4_4V9F_007 | 4V9F | 0.0867 | 0 | 0 | LSU rRNA | G | 1828 | A | 1829 | C | 1830 | U | 1831 | G | 1832 | * | C | 1844 | A | 1845 | U | 1846 | A | 1847 | G | 1848 | * | U | 1883 | G | 1884 | A | 1885 | A | 1886 | U | 1887 | * | A | 2015 | U | 2016 | U | 2017 | A | 2018 | A | 2019 | C | 2020 | cWW | tSW | ntHSa | ntHS | cWW | tHS | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GACUG*CAUAG*UGAUU*AUUAAC | 1 |
| GACUG*CAUAG*UGAAU*AUUAAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| ACU*AUA*GAU*UUAA | 1 |
| ACU*AUA*GAA*UUAA | 1 |
- Annotations
-
- (2)
- Basepair signature
- cWW-tSW-F-F-tHS-F-cWW-F-cWW-F-F-tHS-F-cWW
- Heat map statistics
- Min 0.09 | Avg 0.04 | Max 0.09
Coloring options: