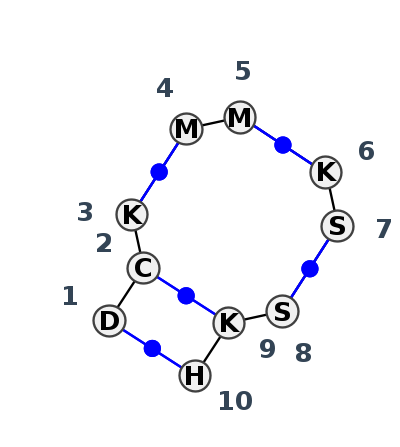
Motif J4_43687.4 Version J4_43687.4 of this group appears in releases 4.1 to 4.1
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | break | 4 | 5 | break | 6 | 7 | break | 8 | 9 | 10 | 1-10 | 2-9 | 3-4 | 5-6 | 7-8 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J4_7VYX_001 | 7VYX | 0.3363 | 0 | C | sgRNA | A | 5 | C | 6 | U | 7 | * | A | 59 | C | 60 | * | G | 82 | C | 83 | * | G | 103 | G | 104 | U | 105 | cWW | ncWW | cWW | cWW | cWW | |
| 2 | J4_7KKV_001 | 7KKV | 0.0000 | 0 | B | OLE* RNA | U | 490 | C | 491 | U | 492 | * | A | 512 | A | 513 | * | U | 526 | G | 527 | * | C | 535 | U | 536 | A | 537 | cWW | cWW | cWW | cWW | cWW | |
| 3 | J4_2A64_001 | 2A64 | 0.5310 | 1 | RNase P junction | A | RNase P class B | G | 136 | C | 138 | G | 139 | * | C | 244 | C | 245 | * | G | 91 | C | 92 | * | G | 113 | G | 114 | C | 115 | cWW | ncWW | cWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| ACU*AC*GC*GGU | 1 |
| UCU*AA*UG*CUA | 1 |
| GC*GGC*GACG*CC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| C***G | 1 |
| C***U | 1 |
| *G*AC* | 1 |
Release history
| Release | 4.1 |
|---|---|
| Date | 2025-09-10 |
| Status | Updated, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (2)
- RNase P junction (1)
- Basepair signature
- cWW-cWW-cWW-cWW-cWW
- Heat map statistics
- Min 0.34 | Avg 0.33 | Max 0.63
Coloring options: