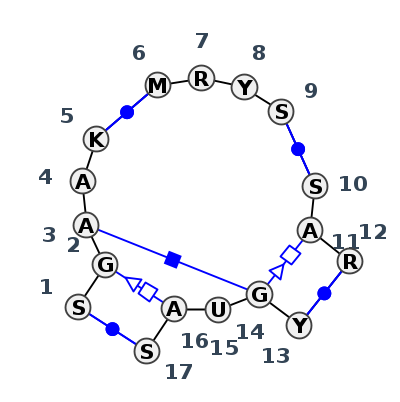
Motif J4_71729.3 Version J4_71729.3 of this group appears in releases 3.54 to 3.86
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | break | 10 | 11 | 12 | break | 13 | 14 | 15 | 16 | 17 | 1-17 | 2-16 | 3-9 | 3-14 | 4-8 | 5-6 | 9-10 | 11-14 | 12-13 | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J4_4WF9_002 | 4WF9 | 0.0912 | 0 | X | LSU rRNA | C | 340 | G | 341 | A | 342 | A | 343 | U | 344 | * | A | 359 | A | 360 | U | 361 | C | 362 | * | G | 366 | A | 367 | A | 368 | * | U | 380 | G | 381 | U | 382 | A | 383 | G | 384 | cWW | cHH | ncSs | cWW | cWW | tHS | cWW | ||
| 2 | J4_7RQB_002 | 7RQB | 0.0000 | 0 | 1A | LSU rRNA | C | 297 | G | 298 | A | 299 | A | 300 | G | 301 | * | C | 316 | G | 317 | C | 318 | C | 319 | * | G | 323 | A | 324 | G | 325 | * | C | 337 | G | 338 | U | 339 | A | 340 | G | 341 | cWW | tSH | cHH | cWW | cWW | tHS | cWW | ||
| 3 | J4_5J7L_019 | 5J7L | 0.0816 | 0 | DA | LSU rRNA | G | 297 | G | 298 | A | 299 | A | 300 | G | 301 | * | C | 316 | G | 317 | C | 318 | G | 319 | * | C | 323 | A | 324 | G | 325 | * | C | 337 | G | 338 | U | 339 | A | 340 | C | 341 | cWW | tSH | cHH | cWW | cWW | tHS | cWW | ||
| 4 | J4_7A0S_002 | 7A0S | 0.0906 | 0 | X | LSU rRNA | C | 308 | G | 309 | A | 310 | A | 311 | G | 312 | * | C | 327 | A | 328 | C | 329 | C | 330 | * | G | 334 | A | 335 | A | 336 | * | U | 348 | G | 349 | U | 350 | A | 351 | G | 352 | cWW | ntSH | ncsS | ncHH | ncSs | cWW | cWW | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGAAU*AAUC*GAA*UGUAG | 1 |
| CGAAG*CGCC*GAG*CGUAG | 1 |
| GGAAG*CGCG*CAG*CGUAC | 1 |
| CGAAG*CACC*GAA*UGUAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAA*GC*A*GUA | 2 |
| GAA*AU*A*GUA | 1 |
| GAA*AC*A*GUA | 1 |
Release history
| Release | 3.54 | 3.55 | 3.56 | 3.57 | 3.58 | 3.59 | 3.60 | 3.61 | 3.62 | 3.63 | 3.64 | 3.65 | 3.66 | 3.67 | 3.68 | 3.69 | 3.70 | 3.71 | 3.72 | 3.73 | 3.74 | 3.75 | 3.76 | 3.77 | 3.78 | 3.79 | 3.80 | 3.81 | 3.82 | 3.83 | 3.84 | 3.85 | 3.86 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Date | 2022-02-02 | 2022-03-02 | 2022-03-30 | 2022-04-27 | 2022-05-25 | 2022-06-22 | 2022-07-20 | 2022-08-17 | 2022-09-14 | 2022-10-12 | 2022-11-09 | 2022-12-07 | 2023-01-04 | 2023-02-01 | 2023-03-01 | 2023-03-29 | 2023-04-26 | 2023-05-24 | 2023-06-21 | 2023-07-19 | 2023-08-16 | 2023-09-13 | 2023-10-11 | 2023-11-08 | 2023-12-06 | 2024-01-03 | 2024-01-31 | 2024-02-28 | 2024-03-27 | 2024-04-24 | 2024-05-22 | 2024-06-19 | 2024-07-17 |
| Status | Updated, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (4)
- Basepair signature
- cWW-tSH-cHH-F-F-tHS-cWW-cWW-F-F-cWW
- Heat map statistics
- Min 0.08 | Avg 0.07 | Max 0.11
Coloring options: