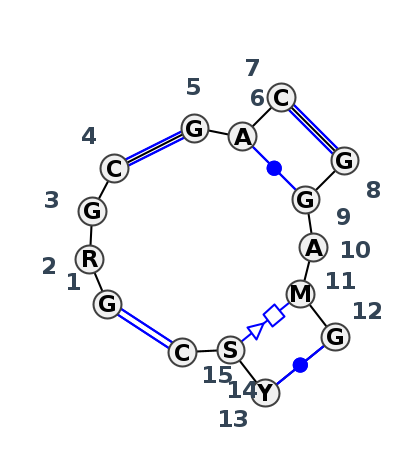
Motif J4_77044.3 Version J4_77044.3 of this group appears in releases 3.87 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | break | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | break | 13 | 14 | 15 | 1-15 | 4-5 | 6-9 | 7-8 | 11-14 | 12-13 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J4_5J7L_028 | 5J7L | 0.1164 | 0 | DA | LSU rRNA | G | 2643 | G | 2644 | G | 2645 | C | 2646 | * | G | 2674 | A | 2675 | C | 2676 | * | G | 2731 | G | 2732 | A | 2733 | A | 2734 | G | 2735 | * | U | 2769 | G | 2770 | C | 2771 | cWW | cWW | cWW | cWW | tHS | cWW |
| 2 | J4_8VTW_010 | 8VTW | 0.0000 | 0 | 1A | LSU rRNA | G | 2643 | G | 2644 | G | 2645 | C | 2646 | * | G | 2674 | A | 2675 | C | 2676 | * | G | 2731 | G | 2732 | A | 2733 | A | 2734 | G | 2735 | * | C | 2769 | G | 2770 | C | 2771 | cWW | cWW | cWW | cWW | tHS | cWW |
| 3 | J4_4WF9_009 | 4WF9 | 0.1449 | 0 | X | LSU rRNA | G | 2670 | A | 2671 | G | 2672 | C | 2673 | * | G | 2701 | A | 2702 | C | 2703 | * | G | 2758 | G | 2759 | A | 2760 | C | 2761 | G | 2762 | * | C | 2796 | C | 2797 | C | 2798 | cWW | cWW | cWW | cWW | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GGGC*GAC*GGAAG*UGC | 1 |
| GGGC*GAC*GGAAG*CGC | 1 |
| GAGC*GAC*GGACG*CCC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GG*A*GAA*G | 2 |
| AG*A*GAC*C | 1 |
- Annotations
-
- (3)
- Basepair signature
- cWW-F-tHS-F-cWW-cWW-cWW-F-cWW
- Heat map statistics
- Min 0.12 | Avg 0.09 | Max 0.16
Coloring options: