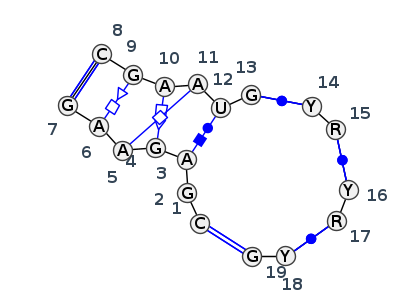
Motif J5_75459.1 Version J5_75459.1 of this group appears in releases 4.0 to 4.7
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 13 | break | 14 | 15 | break | 16 | 17 | break | 18 | 19 | 1-19 | 2-3 | 3-12 | 4-10 | 5-11 | 6-9 | 7-8 | 13-14 | 15-16 | 17-18 | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | J5_9E6Q_001 | 9E6Q | 0.0000 | 1 | 1 | LSU rRNA | C | 39 | G | 40 | A | 41 | G | 43 | A | 44 | A | 45 | G | 46 | * | C | 107 | G | 108 | A | 109 | A | 110 | U | 111 | G | 112 | * | U | 121 | G | 122 | * | C | 144 | G | 145 | * | C | 158 | G | 159 | cWW | ncSs | cHW | tSH | tHH | tHS | cWW | cWW | cWW | cWW |
| 2 | J5_7A0S_001 | 7A0S | 0.1451 | 1 | X | LSU rRNA | C | 46 | G | 47 | A | 48 | G | 50 | A | 51 | A | 52 | G | 53 | * | C | 114 | G | 115 | A | 116 | A | 117 | U | 118 | G | 119 | * | C | 128 | A | 129 | * | U | 142 | A | 143 | * | U | 154 | G | 155 | cWW | ncSs | cHW | tSH | tHH | tHS | cWW | cWW | cWW | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGAAGAAG*CGAAUG*UG*CG*CG | 1 |
| CGAUGAAG*CGAAUG*CA*UA*UG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAAGAA*GAAU*** | 1 |
| GAUGAA*GAAU*** | 1 |
Release history
| Release | 4.0 | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 | 4.7 |
|---|---|---|---|---|---|---|---|---|
| Date | 2025-08-13 | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 | 2026-02-25 |
| Status | New id, 1 parent | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (2)
- Basepair signature
- cWW-F-cWW-cHW-tSH-cWW-tHH-tHS-cWW-cWW
- Heat map statistics
- Min 0.15 | Avg 0.07 | Max 0.15
Coloring options: