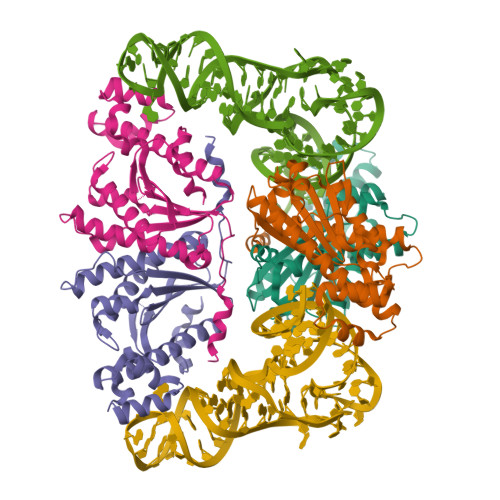
3WC2 Summary
- Structure title
- Crystal structure of C. albicans tRNA(His) guanylyltransferase (Thg1) with a tRNA(Phe)(GUG)
- Authors
- Nakamura, A., Nemoto, T., Sonoda, T., Yamashita, K., Tanaka, I., Yao, M.
- Release date
- 2013-12-18
- Experimental technique
- X-RAY DIFFRACTION
- Resolution
- 3.6 Å
- Chains
- 2 RNA chains from unknown source
- 4 other chains
- Nucleic acid compounds
- Other compounds
- 76mer-tRNA, 76mer-tRNA
- Likely histidyl tRNA-specific guanylyltransferase, Likely histidyl tRNA-specific guanylyltransferase, Likely histidyl tRNA-specific guanylyltransferase, Likely histidyl tRNA-specific guanylyltransferase
Pairwise Interactions
Interactions annotated by FR3D:- 42 base-pairs
- 140 base-stacking
- 4 base-phosphate
- 4 base-ribose
RNA 3D Motifs
In the current release of the RNA 3D Motif Atlas, 3WC2 contains:- 7 hairpin loops from 0 motif groups
- 0 internal loops from 0 motif groups
- 3 junction loops from 0 motif groups
3D Redundancy
3WC2 has chains which belong to the following equivalence classes in the current representative set:- NR_all_83593.11 (2)
- NR_4.0_83593.11 (2)
- NR_20.0_83593.11 (2)
Similar structures
5AXM 1EVV 6XZB 9YPG 9YPO 8CDR 8CEH 6LVR 5AXN 9YPS 5M1J 1OB2 9YPW 9YPY 9YPV 6LVR 9YPT 8CF5 9Y4G 9F9S 6GQV 6XIQ 6GZ3 8CTH 6XIR 6XIQ 6GQB 6GZ5 8CG8 6GQ1 6XIR 4V69 1SZ1 1SZ1 1QZB 1QZA 4TNA 1EHZ 1LS2 8CCS 8CIV 1I9V 1TN1 1TN2 4TRA 6TNA 1TTT 1TTT 1TTT 6XZ7 1JGQ 8CDL 1TRA 8CKU 1JGQ 4V42 1JGO 4V42 1JGO 1JGP 1JGP 1FCW 1FCW 1FCW 1FCW 1FCWStructures from NR_all_83593.11