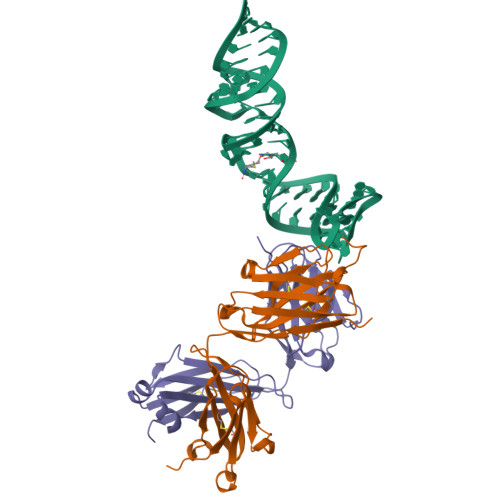
- Structure title
- Crystal structure of a self-alkylating ribozyme - alkylated form with biotinylated epoxide substrate
- Authors
- Koirala, D., Piccirilli, J.A.
- Release date
- 2022-01-19
- Experimental technique
- X-RAY DIFFRACTION
- Resolution
- 1.7 Å
- Chains
-
2 RNA chains from
Aeropyrum pernix
-
4 other chains
- Nucleic acid compounds
Self-alkylating ribozyme (58-MER), Self-alkylating ribozyme (58-MER)
- Other compounds
Fab HAVx Heavy Chain, Fab HAVx Light Chain, Fab HAVx Heavy Chain, Fab HAVx Light Chain
Pairwise Interactions
Interactions annotated by
FR3D:
View all
RNA 3D Motifs
In the current release of the
RNA 3D Motif Atlas, 6XJQ contains:
-
4 hairpin loops from 1 motif group
-
8 internal loops from 4 motif groups
-
0 junction loops from 0 motif groups
More
3D Redundancy
6XJQ has chains which belong to the following equivalence classes in the current representative set:
Similar structures
None found