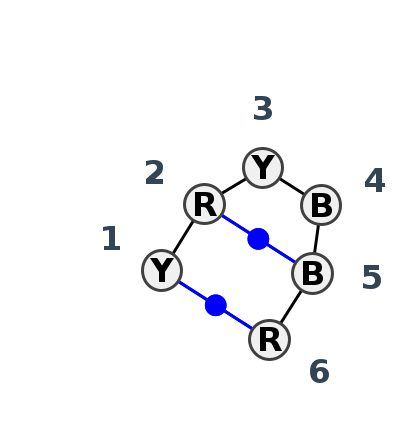
Motif HL_07579.4 Version HL_07579.4 of this group appears in releases 3.88 to 3.88
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 1-6 | 2-5 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_5J7L_213 | 5J7L | 0.7075 | 4 | DA | LSU rRNA | C | 1170 | G | 1171 | C | 1172 | G | 1177 | C | 1178 | G | 1179 | cWW | ncWW | |
| 2 | HL_4JF2_002 | 4JF2 | 0.4190 | 10 | A | PreQ1-II Riboswitch | U | 28 | A | 29 | C | 30 | U | 41 | G | 42 | A | 43 | cWW | cWW | |
| 3 | HL_5D5L_002 | 5D5L | 0.0000 | 10 | A | PreQ1-II riboswitch | U | 28 | A | 29 | C | 30 | U | 41 | G | 42 | A | 43 | cWW | cWW | |
| 4 | HL_8C3A_210 | 8C3A | 0.6417 | 11 | Pseudoknot geometry (H) | CM | SSU rRNA | U | 813 | A | 814 | U | 815 | C | 827 | U | 828 | A | 829 | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| UACUUAUUUCCUUUGA | 2 |
| CGCUUAUGCG | 1 |
| UAUUUUGUUGGUUUCUA | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| ACUUAUUUCCUUUG | 2 |
| GCUUAUGC | 1 |
| AUUUUGUUGGUUUCU | 1 |
Release history
| Release | 3.88 |
|---|---|
| Date | 2024-09-11 |
| Status | Updated, 1 parent |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (3)
- Pseudoknot geometry (H) (1)
- Basepair signature
- cWW-cWW-F-F
- Heat map statistics
- Min 0.42 | Avg 0.48 | Max 0.72
Coloring options: