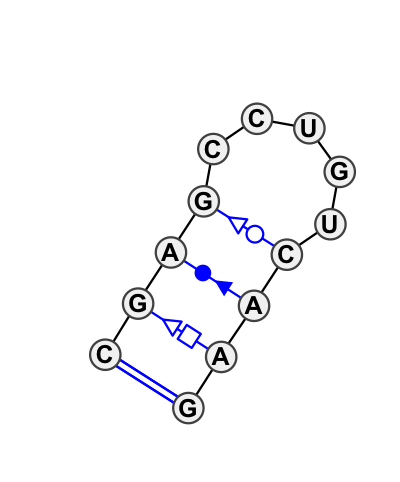
Motif HL_24473.2 Version HL_24473.2 of this group appears in releases 0.2 to 0.2
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 1-13 | 2-12 | 3-11 | 4-10 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_1WZ2_001 | 1WZ2 | 0.5985 | 2 | C | tRNA | C | 912 | G | 913 | A | 914 | G | 915 | C | 916 | C | 917 | U | 918 | G | 920 | U | 921 | C | 922 | A | 924 | A | 925 | G | 926 | cWW | tSH | ncWS | ntSW | |
| 2 | HL_1WZ2_005 | 1WZ2 | 0.0000 | 2 | tRNA D-loop | D | tRNA | C | 912 | G | 913 | A | 914 | G | 915 | C | 916 | C | 917 | U | 918 | G | 920 | U | 921 | C | 922 | A | 924 | A | 925 | G | 926 | cWW | tSH | cWS | tSW |
| 3 | HL_2ZZM_001 | 2ZZM | 0.2814 | 2 | tRNA D-loop | B | tRNA | C | 12 | A | 13 | A | 14 | G | 15 | C | 16 | C | 17 | U | 17|||A | G | 19 | C | 20 | C | 20|||A | A | 21 | A | 22 | G | 23 | cWW | tSH | cWS | tSW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGAGCCUGGUCAAAG | 2 |
| CAAGCCUGGCCAAAG | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GAGCCUGGUCAAA | 2 |
| AAGCCUGGCCAAA | 1 |
Release history
| Release | 0.2 |
|---|---|
| Date | 2011-12-08 |
| Status | Updated, 2 parents |
Parent motifs
| Parent motif | Common motif instances | Only in HL_24473.2 | Only in the parent motif |
|---|---|---|---|
| HL_24473.1 Compare | HL_2ZZM_001, HL_1WZ2_001 | HL_1WZ2_005 | |
| HL_98669.1 Compare | HL_1WZ2_005 | HL_2ZZM_001, HL_1WZ2_001 |
Children motifs
| Child motif | Common motif instances | Only in HL_24473.2 | Only in the child motif |
|---|---|---|---|
| HL_24473.3 Compare | HL_1WZ2_005, HL_2ZZM_001 | HL_1WZ2_001 |
- Annotations
-
- tRNA D-loop (2)
- (1)
- Basepair signature
- Heat map statistics
- Min 0.28 | Avg 0.33 | Max 0.61
Coloring options: