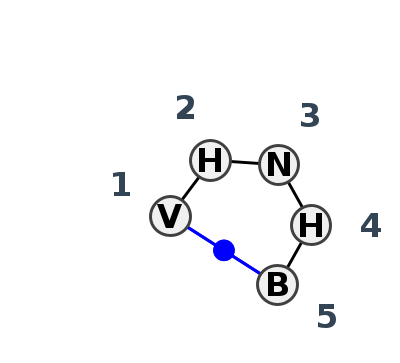
Motif HL_42293.1 Version HL_42293.1 of this group appears in releases 4.1 to 4.7
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 1-5 | 2-4 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_4WF9_048 | 4WF9 | 0.8753 | 2 | X | LSU rRNA | C | 2236 | U | 2237 | A | 2239 | C | 2241 | G | 2242 | ncWW | |
| 2 | HL_8B0X_079 | 8B0X | 0.7955 | 2 | a | LSU rRNA | C | 2213 | U | 2214 | A | 2216 | C | 2218 | G | 2219 | cWW | |
| 3 | HL_2B3J_001 | 2B3J | 0.0000 | 4 | E | anticodon stem-loop of t-RNA-Arg2 (nucleotides 27-42) | A | 31 | C | 32 | G | 36 | A | 38 | U | 39 | cWW | |
| 4 | HL_4V9F_019 | 4V9F | 0.3153 | 2 | 0 | LSU rRNA | G | 670 | A | 671 | U | 673 | U | 675 | C | 676 | cWW | tSH |
| 5 | HL_9E6Q_019 | 9E6Q | 0.3857 | 2 | 1 | LSU rRNA | G | 694 | U | 695 | C | 697 | A | 699 | C | 700 | cWW | ntSH |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CUUAUCG | 1 |
| CUGAUCG | 1 |
| ACUCGGAU | 1 |
| GAGUAUC | 1 |
| GUUCAAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| UUAUC | 1 |
| UGAUC | 1 |
| CU(P5P)CGGA | 1 |
| AGUAU | 1 |
| UUCAA | 1 |
Release history
| Release | 4.1 | 4.2 | 4.3 | 4.4 | 4.5 | 4.6 | 4.7 |
|---|---|---|---|---|---|---|---|
| Date | 2025-09-10 | 2025-10-08 | 2025-11-05 | 2025-12-03 | 2025-12-31 | 2026-01-28 | 2026-02-25 |
| Status | New id, 2 parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (5)
- Basepair signature
- cWW-F-F-F
- Heat map statistics
- Min 0.25 | Avg 0.53 | Max 0.95
Coloring options: