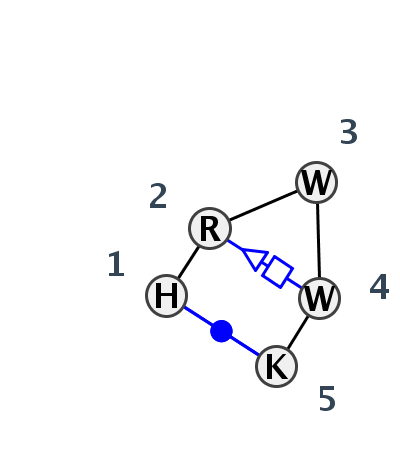
Motif HL_85680.1 Version HL_85680.1 of this group appears in releases 3.77 to 3.87
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 1-5 | 2-4 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | HL_4V9F_007 | 4V9F | 0.0000 | 2 | 0 | LSU rRNA | C | 195 | G | 196 | A | 198 | A | 199 | G | 201 | cWW | tSH |
| 2 | HL_4C9D_002 | 4C9D | 0.2318 | 0 | D | R3 REPEAT RNA CLEAVAGE PRODUCT | C | 21 | G | 22 | U | 23 | A | 24 | G | 25 | ncWW | |
| 3 | HL_1OOA_001 | 1OOA | 0.3575 | 1 | C | RNA aptamer | U | 13 | G | 14 | U | 15 | A | 17 | G | 18 | cWW | ntSH |
| 4 | HL_2DU3_001 | 2DU3 | 0.4926 | 6 | D | tRNA | A | 912 | G | 913 | A | 914 | U | 921 | U | 922 | cWW | ntSH |
| 5 | HL_1U0B_001 | 1U0B | 0.4814 | 6 | A | tRNA | A | 12 | A | 13 | A | 14 | A | 22 | U | 23 | cWW | tSH |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGCAACG | 1 |
| CGUAG | 1 |
| UGUAAG | 1 |
| AGAGGGGCUUU | 1 |
| AAAGCGGUUAU | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GCAAC | 1 |
| GUA | 1 |
| GUAA | 1 |
| GAGGGGCUU | 1 |
| AAGCGGUUA | 1 |
Release history
| Release | 3.77 | 3.78 | 3.79 | 3.80 | 3.81 | 3.82 | 3.83 | 3.84 | 3.85 | 3.86 | 3.87 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Date | 2023-12-06 | 2024-01-03 | 2024-01-03 | 2024-01-31 | 2024-02-28 | 2024-03-27 | 2024-04-24 | 2024-05-22 | 2024-06-19 | 2024-07-17 | 2024-08-14 |
| Status | New id, > 2 parents | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match | Exact match |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- (5)
- Basepair signature
- cWW-tSH-F
- Heat map statistics
- Min 0.23 | Avg 0.35 | Max 0.57
Coloring options: