Motif IL_08257.1 Version IL_08257.1 of this group appears in releases 3.88 to 3.88
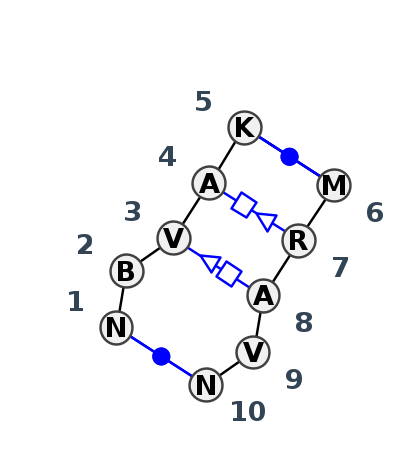
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | break | 6 | 7 | 8 | 9 | 10 | 1-10 | 2-9 | 3-8 | 4-7 | 5-6 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_7A0S_056 | 7A0S | 0.3737 | 0 | Double sheared with near cWW | X | LSU rRNA | C | 1529 | U | 1530 | C | 1531 | A | 1532 | G | 1533 | * | C | 1491 | A | 1492 | A | 1493 | G | 1494 | G | 1495 | cWW | tHS | ncWW | ||
| 2 | IL_7A0S_070 | 7A0S | 0.3619 | 0 | Triple sheared | X | LSU rRNA | U | 1848 | G | 1849 | G | 1850 | A | 1851 | G | 1852 | * | C | 1865 | G | 1866 | A | 1867 | A | 1868 | A | 1869 | cWW | tSH | ntHS | cWW | |
| 3 | IL_2A64_006 | 2A64 | 0.2982 | 0 | Triple sheared | A | RNase P class B | C | 347 | G | 348 | G | 349 | A | 350 | G | 351 | * | C | 378 | G | 379 | A | 380 | A | 381 | G | 382 | cWW | tSH | tSH | cWW | |
| 4 | IL_2A64_005 | 2A64 | 0.2349 | 0 | Triple sheared | A | RNase P class B | C | 297 | G | 298 | G | 299 | A | 300 | G | 301 | * | C | 284 | G | 285 | A | 286 | A | 287 | G | 288 | cWW | tSH | ntSH | tHS | cWW |
| 5 | IL_8C3A_480 | 8C3A | 0.2330 | 0 | Triple sheared | CM | SSU rRNA | C | 1665 | G | 1666 | G | 1667 | A | 1668 | U | 1669 | * | A | 1706 | G | 1707 | A | 1708 | A | 1709 | G | 1710 | cWW | tSH | ntSH | ntWS | cWW |
| 6 | IL_4V88_474 | 4V88 | 0.2219 | 0 | Triple sheared | A6 | SSU rRNA | A | 1678 | G | 1679 | G | 1680 | A | 1681 | U | 1682 | * | A | 1719 | G | 1720 | A | 1721 | A | 1722 | U | 1723 | cWW | tSH | tSH | cWW | |
| 7 | IL_4WFL_001 | 4WFL | 0.0000 | 0 | Double sheared with non-canonical cWW | A | LSRP RNA | U | 72 | C | 73 | G | 74 | A | 75 | G | 76 | * | C | 91 | G | 92 | A | 93 | C | 94 | A | 95 | cWW | cWB | tSH | tHS | cWW |
| 8 | IL_5J7L_274 | 5J7L | 0.1420 | 0 | Double sheared with non-canonical cWW | DA | LSU rRNA | G | 875 | C | 876 | A | 877 | A | 878 | G | 879 | * | C | 898 | A | 899 | A | 900 | C | 901 | C | 902 | cWW | cWB | ntSH | tHS | cWW |
| 9 | IL_5J7L_056 | 5J7L | 0.3361 | 0 | Double sheared with non-canonical cWW | AA | SSU rRNA | G | 1272 | C | 1273 | A | 1274 | A | 1275 | G | 1276 | * | C | 1259 | G | 1260 | A | 1261 | C | 1262 | C | 1263 | cWW | ncWB | tSH | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| CGGAG*CGAAG | 2 |
| CUCAG*CAAGG | 1 |
| UGGAG*CGAAA | 1 |
| CGGAU*AGAAG | 1 |
| AGGAU*AGAAU | 1 |
| UCGAG*CGACA | 1 |
| GCAAG*CAACC | 1 |
| GCAAG*CGACC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| GGA*GAA | 5 |
| UCA*AAG | 1 |
| CGA*GAC | 1 |
| CAA*AAC | 1 |
| CAA*GAC | 1 |
Release history
| Release | 3.88 |
|---|---|
| Date | 2024-09-11 |
| Status | New id, 2 parents |
Parent motifs
This motif has no parent motifs.
Children motifs
This motif has no children motifs.- Annotations
-
- Triple sheared (5)
- Double sheared with non-canonical cWW (3)
- Double sheared with near cWW (1)
- Basepair signature
- cWW-L-R-tSH-tHS-cWW
- Heat map statistics
- Min 0.12 | Avg 0.28 | Max 0.46
Coloring options: