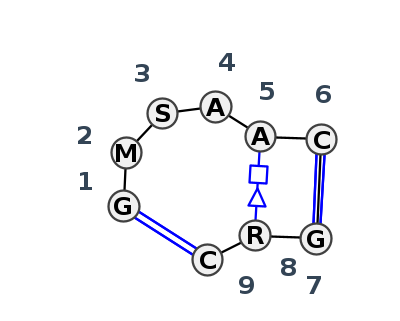
Motif IL_12566.3 Version IL_12566.3 of this group appears in releases 3.87 to 3.90
| #S | Loop id | PDB | Disc | #Non-core | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | break | 7 | 8 | 9 | 1-9 | 2-8 | 5-8 | 6-7 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_4V9F_059 | 4V9F | 0.2281 | 1 | 0 | LSU rRNA | G | 1728 | A | 1729 | C | 1731 | A | 1732 | A | 1733 | C | 1734 | * | G | 2045 | G | 2046 | C | 2047 | cWW | ncWW | tHS | cWW |
| 2 | IL_7A0S_059 | 7A0S | 0.0780 | 0 | X | LSU rRNA | G | 1668 | A | 1669 | G | 1670 | A | 1671 | A | 1672 | C | 1673 | * | G | 1987 | A | 1988 | C | 1989 | cWW | tHS | cWW | |
| 3 | IL_8VTW_063 | 8VTW | 0.0569 | 0 | 1A | LSU rRNA | G | 1651 | A | 1652 | G | 1653 | A | 1654 | A | 1655 | C | 1656 | * | G | 2004 | A | 2005 | C | 2006 | cWW | tHS | cWW | |
| 4 | IL_5J7L_304 | 5J7L | 0.0000 | 0 | DA | LSU rRNA | G | 1651 | A | 1652 | G | 1653 | A | 1654 | A | 1655 | C | 1656 | * | G | 2004 | A | 2005 | C | 2006 | cWW | tHS | cWW | |
| 5 | IL_4WF9_062 | 4WF9 | 0.1134 | 0 | X | LSU rRNA | G | 1695 | C | 1696 | G | 1697 | A | 1698 | A | 1699 | C | 1700 | * | G | 2031 | A | 2032 | C | 2033 | cWW | tHS | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GAGAAC*GAC | 3 |
| GAGCAAC*GGC | 1 |
| GCGAAC*GAC | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| AGAA*A | 3 |
| AGCAA*G | 1 |
| CGAA*A | 1 |
- Annotations
-
- (5)
- Basepair signature
- cWW-L-tHS-L-cWW-L
- Heat map statistics
- Min 0.06 | Avg 0.12 | Max 0.25
Coloring options: