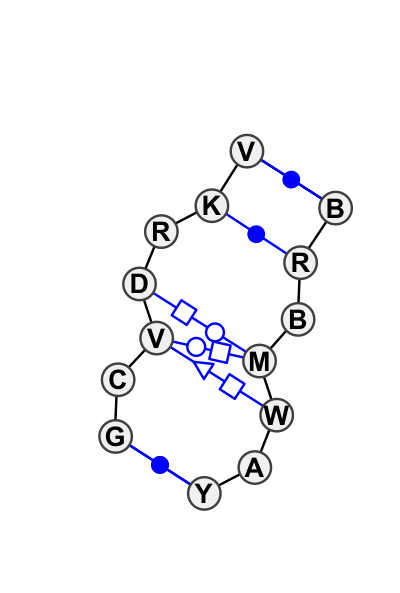
Motif IL_13023.1 Version IL_13023.1 of this group appears in releases 1.17 to 1.17
| #S | Loop id | PDB | Disc | #Non-core | Annotation | Chain(s) | Standardized name for chain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | break | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 1-14 | 2-13 | 3-4 | 3-11 | 3-12 | 4-11 | 5-10 | 6-7 | 6-9 | 7-8 | 9-10 | 12-13 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | IL_1VWW_122 | 1VWW | 0.7455 | 2 | 5 | LSU rRNA | G | 3895 | C | 3896 | G | 3897 | G | 3898 | G | 3899 | G | 3900 | A | 3902 | * | U | 4558 | A | 4559 | C | 4561 | C | 4562 | U | 4563 | A | 4564 | C | 4565 | cWW | ncWW | cSH | cWW | ncSH | ncWH | |||||||
| 2 | IL_3KTW_004 | 3KTW | 0.0000 | 0 | tSH-tHW-tHS with extra cWW pairs | C | SRP RNA | G | 201 | C | 202 | C | 203 | A | 204 | G | 205 | G | 206 | C | 207 | * | G | 216 | A | 217 | G | 218 | C | 219 | A | 220 | A | 221 | C | 222 | cWW | ntWH | tSH | tHW | ntHS | cWW | cWW | |||||
| 3 | IL_1LNG_003 | 1LNG | 0.1502 | 0 | B | SRP S RNA | G | 200 | C | 201 | C | 202 | A | 203 | G | 204 | G | 205 | C | 206 | * | G | 215 | A | 216 | G | 217 | C | 218 | A | 219 | A | 220 | C | 221 | cWW | tSH | tHW | tHS | cWW | cWW | |||||||
| 4 | IL_1VX2_001 | 1VX2 | 0.7414 | 0 | 2 | SSU rRNA | G | 23 | C | 24 | A | 25 | U | 26 | A | 27 | U | 28 | G | 29 | * | C | 645 | G | 646 | U | 647 | A | 648 | U | 649 | A | 650 | U | 651 | cWW | ncWW | cwW | ncWH | cWW |
3D structures
Complete motif including flanking bases
| Sequence | Counts |
|---|---|
| GCCAGGC*GAGCAAC | 2 |
| GCGGGGAA*UACCCUAC | 1 |
| GCAUAUG*CGUAUAU | 1 |
Non-Watson-Crick part of the motif
| Sequence | Counts |
|---|---|
| CCAGG*AGCAA | 2 |
| CGGGGA*ACCCUA | 1 |
| CAUAU*GUAUA | 1 |
Release history
| Release | 1.17 |
|---|---|
| Date | 2014-09-04 |
| Status | New id, 1 parent |
Parent motifs
| Parent motif | Common motif instances | Only in IL_13023.1 | Only in the parent motif |
|---|---|---|---|
| IL_97833.1 Compare | IL_3KTW_004, IL_1LNG_003 | IL_1VWW_122, IL_1VX2_001 |
Children motifs
| Child motif | Common motif instances | Only in IL_13023.1 | Only in the child motif |
|---|---|---|---|
| IL_19589.1 Compare | IL_3KTW_004, IL_1LNG_003 | IL_1VWW_122, IL_1VX2_001 |
- Annotations
-
- (3)
- tSH-tHW-tHS with extra cWW pairs (1)
- Basepair signature
- cWW-L-R-tSH-tWH-tHW-L-R-cWW-cWW
- Heat map statistics
- Min 0.15 | Avg 0.50 | Max 0.85
Coloring options: